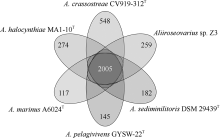
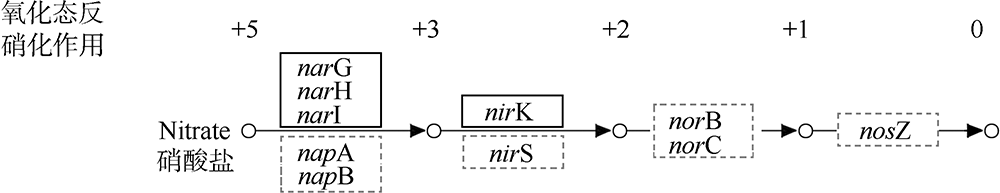
| [1] |
刘志迎, 许海, 詹旭, 等, 2019. 蓝藻水华对太湖水柱反硝化作用的影响[J]. 环境科学, 40(3): 1261-1269.
|
|
LIU ZHIYING, XU HAI, ZHJAN XU, et al, 2019. Influence of Cyanobacterial blooms on denitrification rate in shallow Lake Taihu, China[J]. Environmental Science, 40(3): 1261-1269. (in Chinese with English abstract)
|
| [2] |
ABDELHAMED H, NHO S W, KARSI A, et al, 2021. The role of denitrification genes in anaerobic growth and virulence of Flavobacterium columnare[J]. Journal of Applied Microbiology, 130(4): 1062-1074.
doi: 10.1111/jam.v130.4
|
| [3] |
BOETTCHER K J, BARBER B J, SINGER J T, 1999. Use of antibacterial agents to elucidate the etiology of juvenile oyster disease (JOD) in Crassostrea virginica and numerical dominance of an α-proteobacterium in JOD-affected animals[J]. Applied and Environmental Microbiology, 65(6): 2534-2539.
doi: 10.1128/AEM.65.6.2534-2539.1999
|
| [4] |
BOETTCHER K J, GEAGHAN K K, MALOY A P, et al, 2005. Roseovarius crassostreae sp. nov., a member of the Roseobacter clade and the apparent cause of juvenile oyster disease (JOD) in cultured Eastern oysters[J]. International Journal of Systematic and Evolutionary Microbiology, 55(4): 1531-1537.
doi: 10.1099/ijs.0.63620-0
|
| [5] |
BUCHAN A, GONZALEZ J M, MORAN M A, 2005. Overview of the marine Roseobacter lineage[J]. Applied and Environmental Microbiology, 71(10): 5665-5677.
doi: 10.1128/AEM.71.10.5665-5677.2005
|
| [6] |
CAMARGO J A, ALONSO A, SALAMANCA A, 2005. Nitrate toxicity to aquatic animals: a review with new data for freshwater invertebrates[J]. Chemosphere, 58(9): 1255-1267.
doi: 10.1016/j.chemosphere.2004.10.044
|
| [7] |
CASTRESANA J, 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis[J]. Molecular Biology and Evolution, 17(4): 540-552.
doi: 10.1093/oxfordjournals.molbev.a026334
|
| [8] |
CHOI D H, CHO B C, 2006. Citreimonas salinaria gen. nov., sp. nov., a member of the Roseobacter clade isolated from a solar saltern[J]. International Journal of Systematic and Evolutionary Microbiology, 56(12): 2799-2803.
doi: 10.1099/ijs.0.64373-0
|
| [9] |
DARLING A C E, MAU B, BLATTNER F R, et al, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Research, 14(7): 1394-1403.
doi: 10.1101/gr.2289704
|
| [10] |
EDGAR R C, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Research, 32(5): 1792-1797.
doi: 10.1093/nar/gkh340
|
| [11] |
EMMS D M, KELLY S, 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics[J]. Genome Biology, 20(1): 238.
doi: 10.1186/s13059-019-1832-y
|
| [12] |
GRAHAM D W, TRIPPETT C, DODDS W K, et al, 2010. Correlations between in situ denitrification activity and nir-gene abundances in pristine and impacted prairie streams[J]. Environmental Pollution, 158(10): 3225-3229.
doi: 10.1016/j.envpol.2010.07.010
|
| [13] |
GUO HAIPENG, HUANG LEI, HU SONGTAO, et al, 2020. Effects of carbon/nitrogen ratio on growth, intestinal microbiota and metabolome of shrimp (Litopenaeus vannamei)[J]. Frontiers in Microbiology, 11: 652.
doi: 10.3389/fmicb.2020.00652
|
| [14] |
HUANG LEI, GUO HAIPENG, et al, 2020. The bacteria from large-sized bioflocs are more associated with the shrimp gut microbiota in culture system[J]. Aquaculture, 523: 735159.
doi: 10.1016/j.aquaculture.2020.735159
|
| [15] |
INGLIN R C, MEILE L, STEVENS M J A, 2018. Clustering of pan- and core-genome of Lactobacillus provides novel evolutionary insights for differentiation[J]. BMC Genomics, 19(1): 284.
doi: 10.1186/s12864-018-4601-5
|
| [16] |
JUNG Y T, LEE J S, OH K H, et al, 2011. Roseovarius marinus sp. nov., isolated from seawater[J]. International Journal of Systematic and Evolutionary Microbiology, 61(2): 427-432.
doi: 10.1099/ijs.0.019828-0
|
| [17] |
KANEHISA M, GOTO S, 2000. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research, 28(1): 27-30.
doi: 10.1093/nar/28.1.27
|
| [18] |
KENT A G, GARCIA C A, MARTINY A C, 2018. Increased biofilm formation due to high-temperature adaptation in marine Roseobacter[J]. Nature Microbiology, 3(9): 989-995.
doi: 10.1038/s41564-018-0213-8
|
| [19] |
KESSNER L, 2015. Characterization of biofilm formation, chemotaxis, and the genome of Aliiroseovarius crassostreae[D]. Kingston: University of Rhode Island.
|
| [20] |
KIM M, OH H S, PARK S C, et al, 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes[J]. International Journal of Systematic and Evolutionary Microbiology, 64(Pt 2): 346-351.
doi: 10.1099/ijs.0.059774-0
|
| [21] |
KIM Y O, KONG H J, Park S, et al, 2012. Roseovarius halocynthiae sp. nov., isolated from the sea squirt Halocynthia roretzi[J]. International Journal of Systematic and Evolutionary Microbiology, 62(Pt 4): 931-936.
doi: 10.1099/ijs.0.031674-0
|
| [22] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096
|
| [23] |
LEE I, KIM Y O, PARK S C, et al, 2016. OrthoANI: An improved algorithm and software for calculating average nucleotide identity[J]. International Journal of Systematic and Evolutionary Microbiology, 66(2): 1100-1103.
doi: 10.1099/ijsem.0.000760
|
| [24] |
LI WEIZHONG, GODZIK A, 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences[J]. Bioinformatics, 22(13): 1658-1659.
doi: 10.1093/bioinformatics/btl158
|
| [25] |
LUO HAIWEI, LÖYTYNOJA A, MORAN M A, 2012. Genome content of uncultivated marine Roseobacters in the surface ocean[J]. Environmental Microbiology, 14(1): 41-51.
doi: 10.1111/j.1462-2920.2011.02528.x
|
| [26] |
MASELLA A P, BARTRAM A K, TRUSZKOWSKI J M, et al, 2012. PANDAseq: paired-end assembler for illumina sequences[J]. BMC Bioinformatics, 13: 31.
doi: 10.1186/1471-2105-13-31
|
| [27] |
MEIER-KOLTHOFF J P, AUCH A F, KLENK H P, et al, 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions[J]. BMC Bioinformatics, 14(1): 60.
doi: 10.1186/1471-2105-14-60
|
| [28] |
MOU XIAOZHEN, SUN SHULEI, EDWARDS R A, et al, 2008. Bacterial carbon processing by generalist species in the coastal ocean[J]. Nature, 451(7179): 708-711.
doi: 10.1038/nature06513
|
| [29] |
NATALE D A, SHANKAVARAM U T, GALPERIN M Y, et al, 2000. Towards understanding the first genome of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs)[J]. Genome Biology, 1(5): RESEARCH0009.
|
| [30] |
PARK S, PARK J M, KANG C H, et al, 2015. Aliiroseovarius pelagivivens gen. nov., sp. nov., isolated from seawater, and reclassification of three species of the genus Roseovarius as Aliiroseovarius crassostreae comb. nov., Aliiroseovarius halocynthiae comb. nov. and Aliiroseovarius sediminilitoris comb. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 65(Pt 8): 2646-2652.
doi: 10.1099/ijs.0.000315
|
| [31] |
PARK S, YOON J H, 2013. Roseovarius sediminilitoris sp. nov., isolated from seashore sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 63(5): 1741-1745.
doi: 10.1099/ijs.0.043737-0
|
| [32] |
PHILIPPOT L, 2002. Denitrifying genes in bacterial and Archaeal genomes[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1577(3): 355-376.
doi: 10.1016/S0167-4781(02)00420-7
|
| [33] |
PUJALTE M J, LUCENA T, RUVIRA M A, et al, 2014. The family Rhodobacteraceae[M]// The Prokaryotes. Berlin, Heidelberg: Springer: 439-512.
|
| [34] |
REISCH C R, MORAN M A, WHITMAN W B, 2011. Bacterial catabolism of dimethylsulfoniopropionate (DMSP)[J]. Frontiers in Microbiology, 2: 172.
|
| [35] |
RICHTER M, ROSSELLÓ-MÓRA R, 2009. Shifting the genomic gold standard for the prokaryotic species definition[J]. Proceedings of the National Academy of Sciences of the United States of America, 106(45): 19126-19131.
|
| [36] |
SHARPE G C, GIFFORD S M, SEPTER A N, 2020. A model Roseobacter, Ruegeria pomeroyi DSS-3, Employs a diffusible killing mechanism to eliminate competitors[J]. mSystems, 5(4): e00443-20.
|
| [37] |
STAMATAKIS A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 30(9): 1312-1313.
doi: 10.1093/bioinformatics/btu033
|
| [38] |
SUN SHENGMING, GE XIANPING, ZHU JIAN, et al, 2014. Identification and mRNA expression of antioxidant enzyme genes associated with the oxidative stress response in the Wuchang bream (Megalobrama amblycephala Yih) in response to acute nitrite exposure[J]. Comparative Biochemistry and Physiology Part C: Toxicology and Pharmacology, 159: 69-77.
doi: 10.1016/j.cbpc.2013.09.005
|
| [39] |
XING WEI, LI DESHENG, LI JINLONG, et al, 2016. Nitrate removal and microbial analysis by combined micro- electrolysis and autotrophic denitrification[J]. Bioresource Technology, 211: 240-247.
doi: 10.1016/j.biortech.2016.03.044
|
| [40] |
YU ZEHUI, GENG YI, WANG KAIYU, et al, 2017. Complete genome sequence of Vibrio mimicus strain SCCF01 with potential application in fish vaccine development[J]. Virulence, 8(6): 1028-1030.
doi: 10.1080/21505594.2016.1250996
|
| [41] |
ZEB S, GULFAM S M, BOKHARI H, 2020. Comparative core/pan genome analysis of Vibrio cholerae isolates from Pakistan[J]. Infection, Genetics and Evolution, 82: 104316.
doi: 10.1016/j.meegid.2020.104316
|
| [42] |
ZHANG XIAN, LIU ZHENGHUA, WEI GUANYUN, et al, 2018. In silico genome-wide analysis reveals the potential links between core genome of Acidithiobacillus thiooxidans and its autotrophic lifestyle[J]. Frontiers in Microbiology, 9: 1255.
doi: 10.3389/fmicb.2018.01255
|
 ), 郭海朋1,2(
), 郭海朋1,2( ), 董鹏生1,2, 燕孟琛1,2, 张德民1,2
), 董鹏生1,2, 燕孟琛1,2, 张德民1,2
 ), GUO Haipeng1,2(
), GUO Haipeng1,2( ), DONG Pengsheng1,2, YAN Mengchen1,2, ZHANG Demin1,2
), DONG Pengsheng1,2, YAN Mengchen1,2, ZHANG Demin1,2