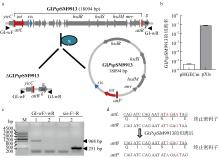
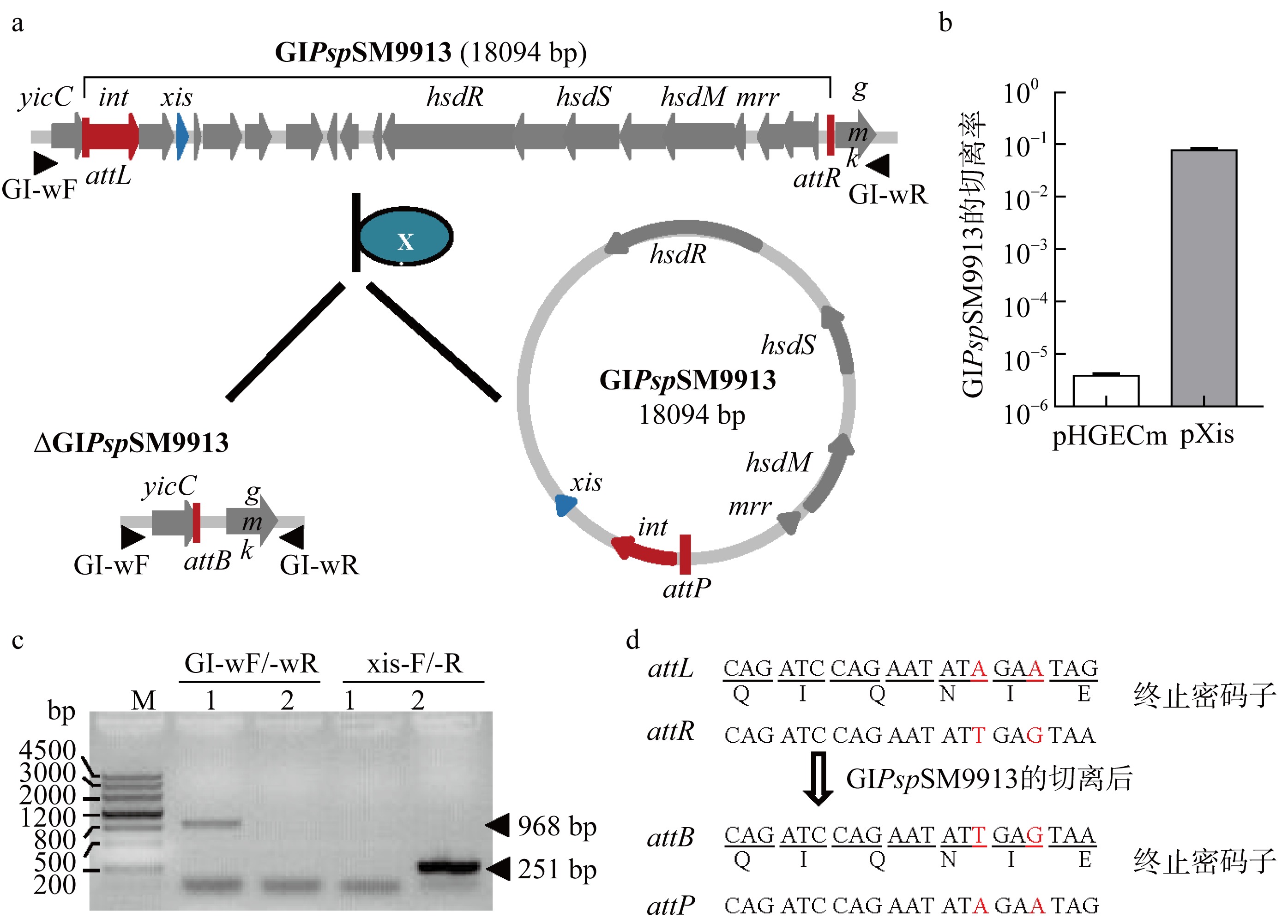
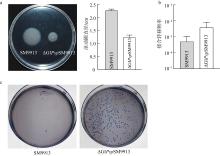
| [1] |
BIAN FEI, XIE BINBIN, QIN QILONG, et al, 2012. Genome sequences of six Pseudoalteromonas strains isolated from Arctic sea ice[J]. Journal of Bacteriology, 194(4): 908-909.
doi: 10.1128/JB.06427-11
|
| [2] |
CASJENS S, 2003. Prophages and bacterial genomics: what have we learned so far?[J]. Molecular Microbiology, 49(2): 277-300.
doi: 10.1046/j.1365-2958.2003.03580.x
pmid: 12886937
|
| [3] |
DACCORD A, CECCARELLI D, RODRIGUE S, et al, 2013. Comparative analysis of mobilizable genomic islands[J]. Journal of Bacteriology, 195(3): 606-614.
doi: 10.1128/JB.01985-12
pmid: 23204461
|
| [4] |
DARLING A C, MAU B, BLATTNER F R, et al, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Research, 14(7): 1394-1403.
doi: 10.1101/gr.2289704
pmid: 15231754
|
| [5] |
FISCHER W, WINDHAGER L, ROHRER S, et al, 2010. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type Ⅳ secretion system and genomic island transfer[J]. Nucleic Acids Research, 38(18): 6089-6101.
doi: 10.1093/nar/gkq378
|
| [6] |
GAILLARD M, PERNET N, VOGNE C, et al, 2008. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1[J]. Proceedings of the National Academy of Sciences, 105(19): 7058-7063.
doi: 10.1073/pnas.0801269105
|
| [7] |
HEMME C L, GREEN S J, RISHISHWAR L, et al, 2016. Lateral gene transfer in a heavy metal-contaminated-groundwater microbial community[J]. mBio, 7(2): e02234-15.
|
| [8] |
JUHAS M, VAN DER MEER J R, GAILLARD M, et al, 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution[J]. Fems Microbiology Reviews, 33(2): 376-393.
doi: 10.1111/j.1574-6976.2008.00136.x
pmid: 19178566
|
| [9] |
LEE C A, AUCHTUNG J M, MONSON R E, et al, 2007. Identification and characterization of int (integrase), xis (excisionase) and chromosomal attachment sites of the integrative and conjugative element ICEBs1 of Bacillus subtilis[J]. Molecular Microbiology, 66(6): 1356-1369.
|
| [10] |
LI M, KOTETISHVILI M, CHEN Y, et al, 2003. Comparative genomic analyses of the vibrio pathogenicity island and cholera toxin prophage regions in nonepidemic serogroup strains of Vibrio cholerae[J]. Applied and Environmental Microbiology, 69(3): 1728-1738.
doi: 10.1128/AEM.69.3.1728-1738.2003
|
| [11] |
NI SONGWEI, LI BAIYUAN, TANG KAIHAO, et al, 2021. Conjugative plasmid-encoded toxin-antitoxin system PrpT/PrpA directly controls plasmid copy number[J]. Proceedings of the National Academy of Sciences, 118(4): e2011577118.
doi: 10.1073/pnas.2011577118
|
| [12] |
NOTO M J, KREISWIRTH B N, MONK A B, et al, 2008. Gene acquisition at the insertion site for SCCmec, the genomic island conferring methicillin resistance in Staphylococcus aureus[J]. Journal of Bacteriology, 190(4): 1276-1283.
doi: 10.1128/JB.01128-07
|
| [13] |
PENG LIHUA, LIANG XIAO, XU JIAKANG, et al, 2020. Monospecific biofilms of Pseudoalteromonas promote larval settlement and metamorphosis of Mytilus coruscus[J]. Scientific Reports, 10(1): 2577.
doi: 10.1038/s41598-020-59506-1
|
| [14] |
POULIN-LAPRADE D, MATTEAU D, JACQUES P E, et al, 2015. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon[J]. Nucleic Acids Research, 43(4): 2045-2056.
doi: 10.1093/nar/gkv071
|
| [15] |
QIN GUOKUI, ZHU LIZHI, CHEN XIULAN, et al, 2007. Structural characterization and ecological roles of a novel exopolysaccharide from the deep-sea psychrotolerant bacterium Pseudoalteromonas sp. SM9913[J]. Microbiology (Reading), 153(Pt 5): 1566-1572.
doi: 10.1099/mic.0.2006/003327-0
|
| [16] |
RAO D, SKOVHUS T, TUJULA N, et al, 2010. Ability of Pseudoalteromonas tunicata to colonize natural biofilms and its effect on microbial community structure[J]. FEMS Microbiology Ecology, 73(3): 450-457.
|
| [17] |
RAY M D, BOUNDY S, ARCHER G L, et al, 2016. Transfer of the methicillin resistance genomic island among staphylococci by conjugation[J]. Molecular Microbiology, 100(4): 675-685.
doi: 10.1111/mmi.13340
pmid: 26822382
|
| [18] |
SCHMIDT H, HENSEL M, 2004. Pathogenicity islands in bacterial pathogenesis[J]. Clinical Microbiology Reviews, 17(1): 14-56.
doi: 10.1128/CMR.17.1.14-56.2004
pmid: 14726454
|
| [19] |
VOS M, HESSELMAN M C, TE BEEK T A, et al, 2015. Rates of lateral gene transfer in prokaryotes: high but why?[J]. Trends in Microbiology, 23(10): 598-605.
doi: S0966-842X(15)00155-9
pmid: 26433693
|
| [20] |
WADHWA N, BERG H C, 2022. Bacterial motility: machinery and mechanisms[J]. Nat Rev Microbiol, 20(3): 161-173.
doi: 10.1038/s41579-021-00626-4
|
| [21] |
WANG PENGXIA, YU ZICHAO, LI BAIYUAN, et al, 2015. Development of an efficient conjugation-based genetic manipulation system for Pseudoalteromonas[J]. Microbial Cell Factories, 14: 11.
doi: 10.1186/s12934-015-0194-8
|
| [22] |
ZENG ZHENSHUN, GUO XINGPAN, CAI XINGSHENG, et al, 2017. Pyomelanin from Pseudoalteromonas lipolytica reduces biofouling[J]. Microbial Biotechnology, 10(6): 1718-1731.
doi: 10.1111/mbt2.2017.10.issue-6
|
| [23] |
ZHAO DIANLI, YU ZICHAO, LI PINGYI, et al, 2011. Characterization of a cryptic plasmid pSM429 and its application for heterologous expression in psychrophilic Pseudoalteromonas[J]. Microbial Cell Factories, 10: 30.
doi: 10.1186/1475-2859-10-30
|
| [24] |
ZHAO YI, WANG WEIQUAN, YAO JIANJUN, et al, 2022. The HipAB toxin-antitoxin system stabilizes a composite genomic island in Shewanella putrefaciens CN-32[J]. Frontiers in Microbiology, 13: 858857.
doi: 10.3389/fmicb.2022.858857
|
| [25] |
ZHENG YU, COHEN-KARNI D, XU D, et al, 2010. A unique family of Mrr-like modification-dependent restriction endonucleases[J]. Nucleic Acids Research, 38(16): 5527-5534.
doi: 10.1093/nar/gkq327
pmid: 20444879
|
 ), 赵逸5, 杜晓飞1,3, 王伟权1,3, 王晓雪1,2,3
), 赵逸5, 杜晓飞1,3, 王伟权1,3, 王晓雪1,2,3
 ), ZHAO Yi5, DU Xiaofei1,3, WANG Weiquan1,3, WANG Xiaoxue1,2,3
), ZHAO Yi5, DU Xiaofei1,3, WANG Weiquan1,3, WANG Xiaoxue1,2,3