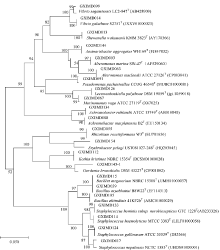
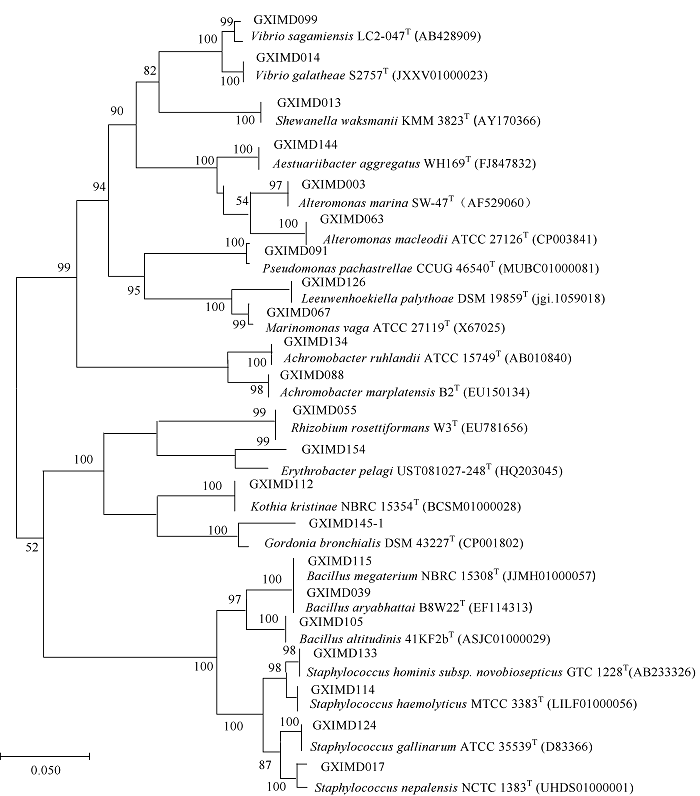
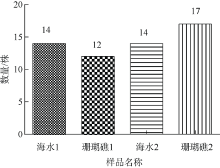
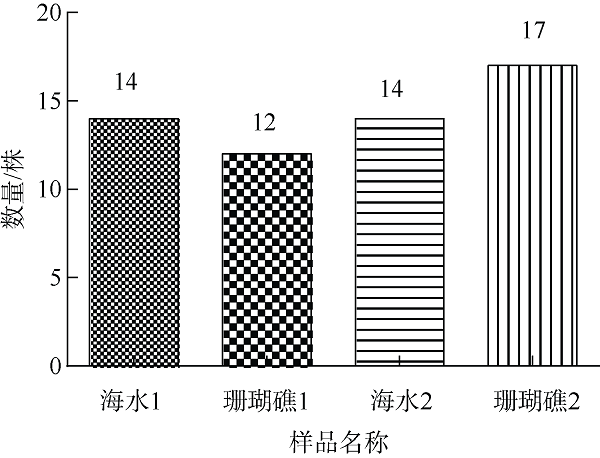
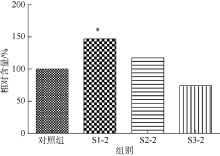
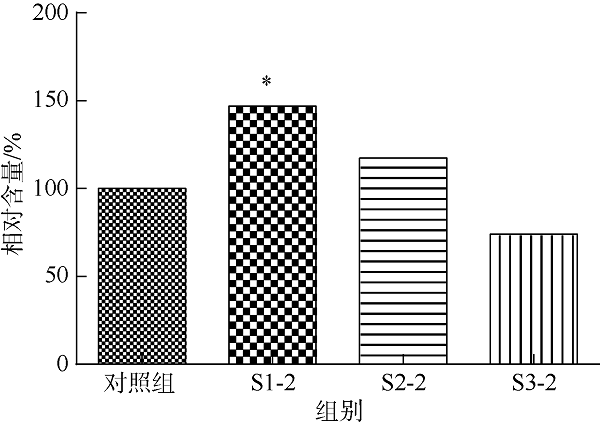
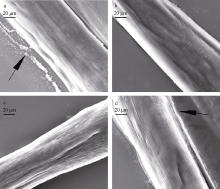
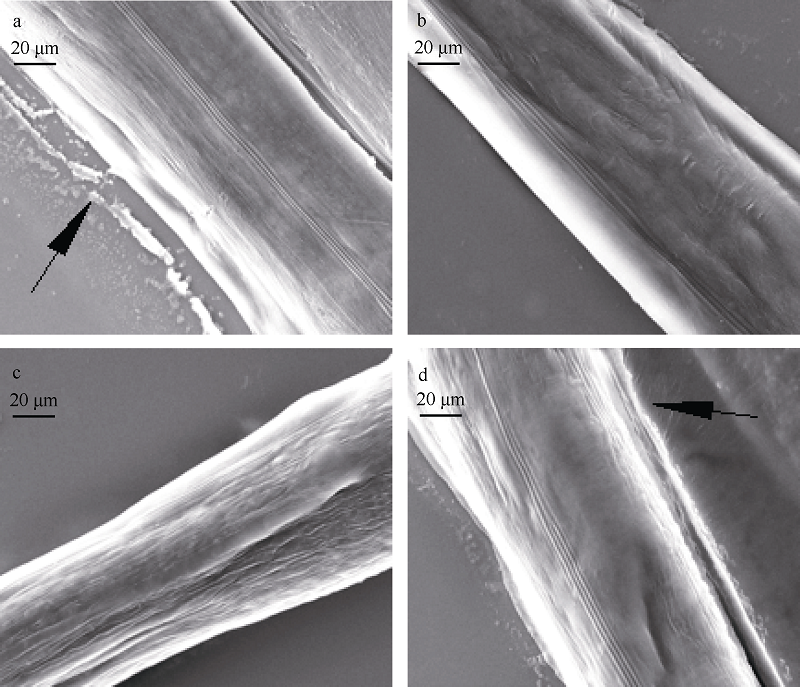
| [1] |
蔡雪芹, 2018. 浅谈珊瑚及其生态功能与保护[J]. 海洋与渔业, (7):100-102 (in Chinese).
|
| [2] |
李菲, 高程海, 余炼, 等, 2018. 川蔓藻内生及根际细菌多样性与抑菌活性研究[J]. 广西植物, 38(7):922-933.
|
|
LI FEI, GAO CHENGHAI, YU LIAN, et al, 2018. Diversity and antifungal activity of endophytic and rhizospheric bacteria isolated from Ruppia maritima[J]. Guihaia, 38(7):922-933 (in Chinese with English abstract).
|
| [3] |
孙静, 王素英, 张德超, 等, 2014. 海南红树林根系土壤中可培养细菌的多样性分析[J]. 海洋科学, 38(7):27-33.
|
|
SUN JING, WANG SUYING, ZHANG DECHAO, et al, 2014. Diversity of culturable bacteria from the soil of root system of mangrove forest of Beigang island in Hainan Province[J]. Marine Sciences, 38(7):27-33 (in Chinese with English abstract).
|
| [4] |
王怀玲, 2018. 蓝莓多酚化合物抗衰老活性及作用机制研究[D]. 广州: 华南理工大学: 1-172.
|
|
WANG HUAILING, 2018. Research on Antiaging activity and mechanism of blueberry polyphenols[D]. Guangzhou: South China University of Technology: 1-172 (in Chinese with English abstract).
|
| [5] |
周双清, 黄小龙, 黄东益, 等, 2010. Chelex-100快速提取放线菌DNA作为PCR扩增模板[J]. 生物技术通报, 22(2):123-125.
|
|
ZHOU SHUANGQIN, HUANG XIAOLONG, HUANG DONGYI, et al, 2010. A rapid method for extracting DNA from actinomycetes by chelex-100[J]. Biotechnology Bulletin, 22(2):123-125 (in Chinese with English abstract).
|
| [6] |
BRENNER S, 1974. The genetics of Caenorhabditis elegans[J]. Genetics, 77(1):71-94.
pmid: 4366476
|
| [7] |
CHISHOLM A D, XU SUHONG, 2012. The Caenorhabditis elegans epidermis as a model skin. II: Differentiation and physiological roles[J]. Wiley Interdisciplinary Reviews: Developmental Biology, 1(6):879-902.
doi: 10.1002/wdev.77
pmid: 23539358
|
| [8] |
HARTMANN A C, PETRAS D, QUINN R A, et al, 2017. Meta-mass shift chemical profiling of metabolomes from coral reefs[J]. Proceedings of the National Academy of Sciences of the United States of America, 114(44):11685-11690.
doi: 10.1073/pnas.1710248114
pmid: 29078340
|
| [9] |
HERNDON L A, SCHMEISSNER P J, DUDARONEK J M, et al, 2002. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans[J]. Nature, 419(6909):808-814.
pmid: 12397350
|
| [10] |
KIM K H, ROH S W, CHANG HW, et al, 2009. Nitratireductor basaltis sp. nov., isolated from black beach sand[J]. International Journal of Systematic and Evolutionary Microbiology, 59(1):135-138.
|
| [11] |
KIM M, OH H S, PARK S C, et al, 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes[J]. International Journal of Systematic and Evolutionary Microbiology, 64(2):346-351.
|
| [12] |
LUEPONGPATTANA S, THANIYAVARN J, MORIKAWA M, 2017. Production of Massoia Lactone by Aureobasidium pullulans YTP6-14 isolated from the gulf of Thailand and its fragrant biosurfactant properties[J]. Journal of Applied Microbiology, 123(6):1488-1497.
doi: 10.1111/jam.13598
pmid: 28972680
|
| [13] |
PARK H E H, JUNG Y, LEE S J V, 2017. Survival assays using Caenorhabditis elegans[J]. Molecules and Cells, 40(2):90-99.
pmid: 28241407
|
| [14] |
SUGI T, 2016. Genome editing in C. elegans and other nematode species[J]. International Journal of Molecular Sciences, 17(3):295.
doi: 10.3390/ijms17030295
pmid: 26927083
|
| [15] |
SUPONG K, SURIYACHADKUN C, PITTAYAKHAJONWUT P, et al, 2013. Micromonospora spongicola sp. nov., an actinomycete isolated from a marine sponge in the Gulf of Thailand[J]. The Journal of Antibiotics, 66(9):505-509.
pmid: 23677031
|
| [16] |
THAWORNWIRIYANUN P, TANASUPAWAT S, DECHSAKUL WATANAC, et al, 2012. Identification of Newly Zeaxanthin- producing bacteria isolated from sponges in the gulf of Thailand and their Zeaxanthin production[J]. Applied Biochemistry and Biotechnology, 167(8):2357-2368.
doi: 10.1007/s12010-012-9760-2
pmid: 22715027
|
 ), 李蜜1, 刘昕明2, 刘永宏3, 龙超3, 钟振国1, 易湘茜1, 高程海1(
), 李蜜1, 刘昕明2, 刘永宏3, 龙超3, 钟振国1, 易湘茜1, 高程海1( )
)
 ), LI Mi1, LIU Xinming2, LIU Yonghong3, LONG Chao3, ZHONG Zhenguo1, YI Xiangxi1, GAO Chenghai1(
), LI Mi1, LIU Xinming2, LIU Yonghong3, LONG Chao3, ZHONG Zhenguo1, YI Xiangxi1, GAO Chenghai1( )
)