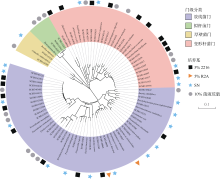
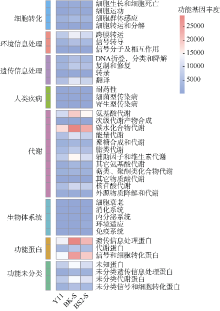
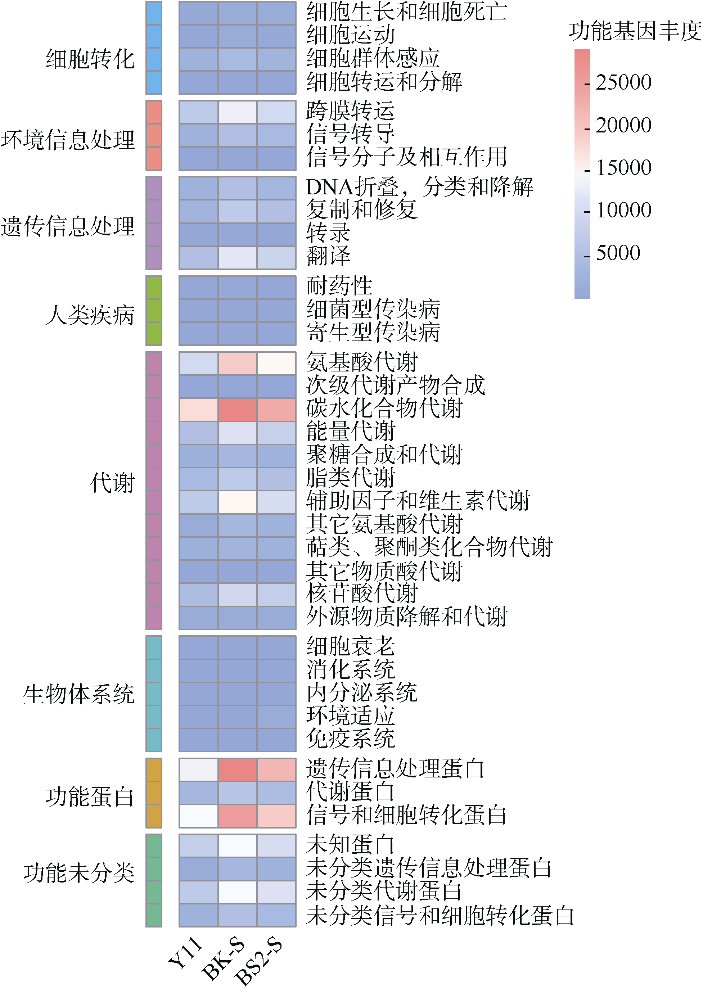
| [1] |
韩敏敏, 李蜜, 刘昕明, 等, 2020. Khai岛和Pathiu岛珊瑚礁沉积物细菌多样性及细菌粗提物延缓秀丽隐杆线虫衰老活性研究[J]. 热带海洋学报, 39(5): 19-29.
|
|
HAN MINMIN, LI MI, LIU XINMING, et al, 2020. Studies on bacterial diversity in coral reef sediments in Khai Island and Pathiu Island and bacterial crude extract retards aging activity of Caenorhabditis elegans[J]. Journal of Tropical Oceanography, 39(5): 19-29. (in Chinese with English abstract)
|
| [2] |
何媛秋, 李存, 陈柔雯, 等, 2020. 不同培养条件对海洋沉积环境细菌的选择性分离[J]. 生物资源, 42(5): 540-548.
|
|
HE YUANQIU, LI CUN, CHEN ROUWEN, et al, 2020. Selective isolation of bacteria from marine sedimentary environment by different culture conditions[J]. Biotic Resources, 42(5): 540-548. (in Chinese with English abstract)
|
| [3] |
龙丽娟, 杨芳芳, 韦章良, 2019. 珊瑚礁生态系统修复研究进展[J]. 热带海洋学报, 38(6): 1-8.
|
|
LONG LIJUAN, YANG FANGFANG, WEI ZHANGLIANG, 2019. A review on ecological restoration techniques of coral reefs[J]. Journal of Tropical Oceanography, 38(6): 1-8. (in Chinese with English abstract)
|
| [4] |
孙创, 王金燕, 张钰琳, 等, 2021. 利用改良培养基探究西太平洋海水可培养细菌多样性[J]. 微生物学报, 61(4): 845-861.
|
|
SUN CHUANG, WANG JINYAN, ZHANG YULIN, et al, 2021. Exploring the diversity of cultivated bacteria in the Western Pacific waters through improved culture media[J]. Acta Microbiologica Sinica, 61(4): 845-861. (in Chinese with English abstract)
|
| [5] |
熊盈盈, 莫祯妮,邱树毅, 等, 2021. 未培养环境微生物培养方法的研究进展[J]. 微生物学通报, 48(5): 1765-1779.
|
|
XIONG YINGYING, MO ZHENNI, QIU SHUYI, et al, 2021. Research progress on culture methods of uncultured environmental microorganisms[J]. Microbiology China, 48(5): 1765-1779(in Chinese with English abstract).
|
| [6] |
周进, 晋慧, 蔡中华, 2014. 微生物在珊瑚礁生态系统中的作用与功能[J]. 应用生态学报, 25(3): 919-930.
|
|
ZHOU JIN, JIN HUI, CAI ZHONGHUA, 2014. A review of the role and function of microbes in coral reef ecosystem[J]. Chinese Journal of Applied Ecology, 25(3): 919-930. (in Chinese with English abstract)
|
| [7] |
CHO J C, GIOVANNONI S J, 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria[J]. Applied and Environmental Microbiology, 70(1): 432-440.
doi: 10.1128/AEM.70.1.432-440.2004
|
| [8] |
DOUGLAS G M, MAFFEI V J, ZANEVELD J R, et al, 2020. PICRUSt2 for prediction of metagenome functions[J]. Nature Biotechnology, 38(6): 685-688.
doi: 10.1038/s41587-020-0548-6
|
| [9] |
ISHIDA Y, KADOTA H, 1981. Growth patterns and substrate requirements of naturally occurring obligate oligotrophs[J]. Microbial Ecology, 7(2): 123-130.
doi: 10.1007/BF02032494
|
| [10] |
JANNASCH H W, JONES G E, 1959. Bacterial populations in sea water as determined by different methods of enumeration[J]. Limnology and Oceanography, 4(2): 128-139.
doi: 10.4319/lo.1959.4.2.0128
|
| [11] |
KJELLEBERG S, MARSHALL K C, HERMANSSON M, 1985. Oligotrophic and copiotrophic marine bacteria-observations related to attachment[J]. FEMS Microbiology Letters, 31(2): 89-96.
|
| [12] |
KOCH A L, 2001. Oligotrophs versus copiotrophs[J]. BioEssays, 23(7): 657-661.
doi: 10.1002/(ISSN)1521-1878
|
| [13] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096
|
| [14] |
LETUNIC I, BORK P, 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments[J]. Nucleic Acids Research, 47(W1): W256-W259.
|
| [15] |
SAITOU N, NEI M, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Molecular Biology and Evolution, 4(4): 406-425.
|
| [16] |
SCHÖTTNER S, PFITZNER B, GRÜNKE S, et al, 2011. Drivers of bacterial diversity dynamics in permeable carbonate and silicate coral reef sands from the Red Sea[J]. Environmental Microbiology, 13(7): 1815-1826.
doi: 10.1111/j.1462-2920.2011.02494.x
|
| [17] |
SWINDELL S R, PLASTERER T N, 1997. SEQMAN[M]// SWINDELL S R. Sequence data analysis guidebook. Methods in molecular medicine™, volume 70. Totowa, NJ: Springer: 75-89.
|
| [18] |
WALSH P S, METZGER D A, HIGUCHI R, 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material[J]. Biotechniques, 10(4): 506-513.
|
| [19] |
XIAN WENDONG, SALAM N, LI MENGMENG, et al, 2020. Network-directed efficient isolation of previously uncultivated Chloroflexi and related bacteria in hot spring microbial mats[J]. NPJ Biofilms and Microbiomes, 6: 20.
doi: 10.1038/s41522-020-0131-4
|
| [20] |
YARZA P, YILMAZ P, PRUESSE E, et al, 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences[J]. Nature Reviews Microbiology, 12(9): 635-645.
doi: 10.1038/nrmicro3330
|
| [21] |
YOON S H, HA S M, KWON S, et al, 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies[J]. International Journal of Systematic and Evolutionary Microbiology, 67(5): 1613-1617.
doi: 10.1099/ijsem.0.001755
|
 ), 崔林青1,3,4, 杨红强2, 龙丽娟1,3(
), 崔林青1,3,4, 杨红强2, 龙丽娟1,3( ), 田新朋1,3(
), 田新朋1,3( )
)
 ), CUI Linqing1,3,4, YANG Hongqiang2, LONG Lijuan1,3(
), CUI Linqing1,3,4, YANG Hongqiang2, LONG Lijuan1,3( ), TIAN Xinpeng1,3(
), TIAN Xinpeng1,3( )
)