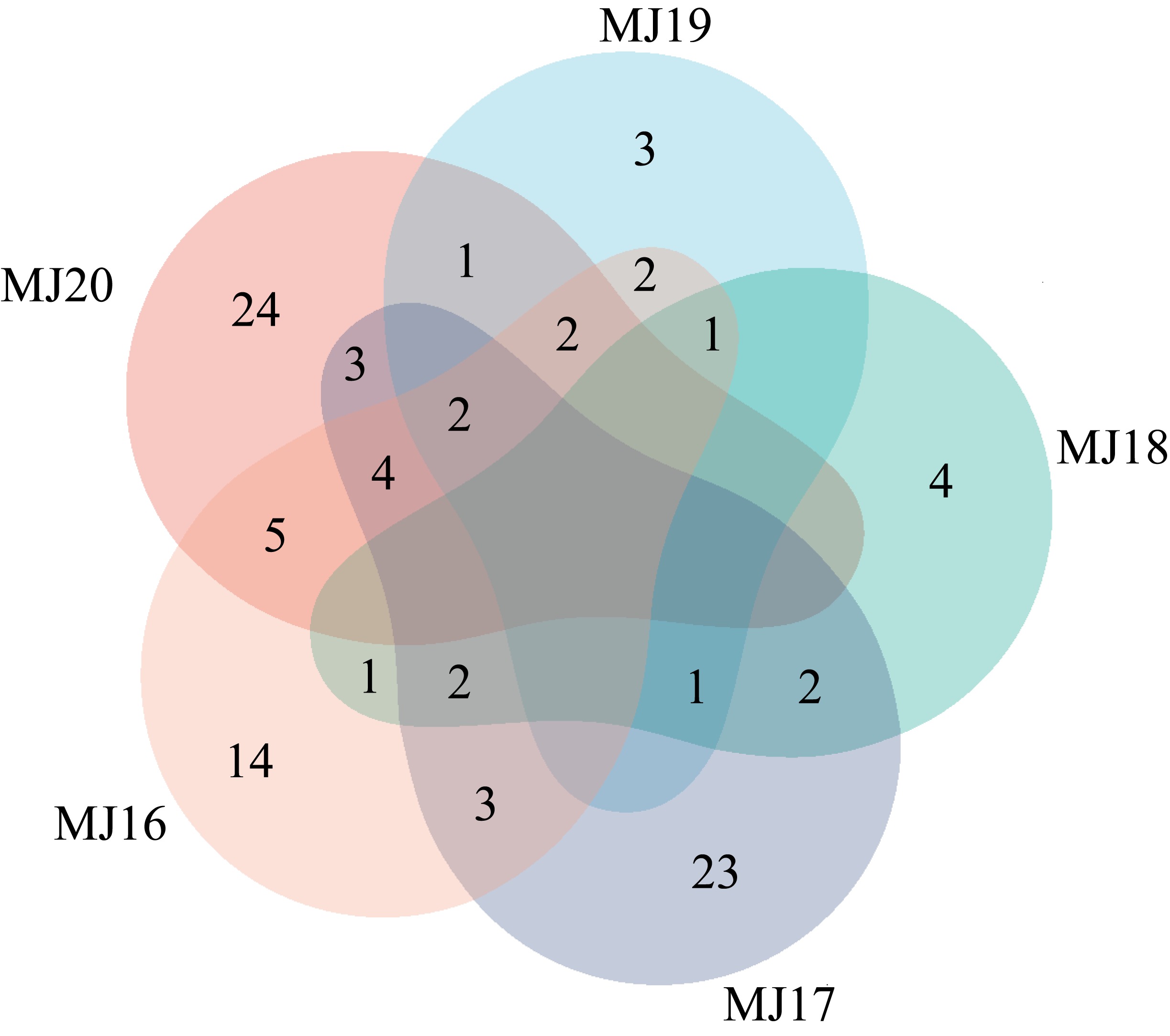
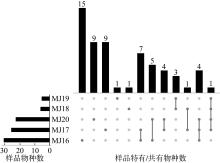
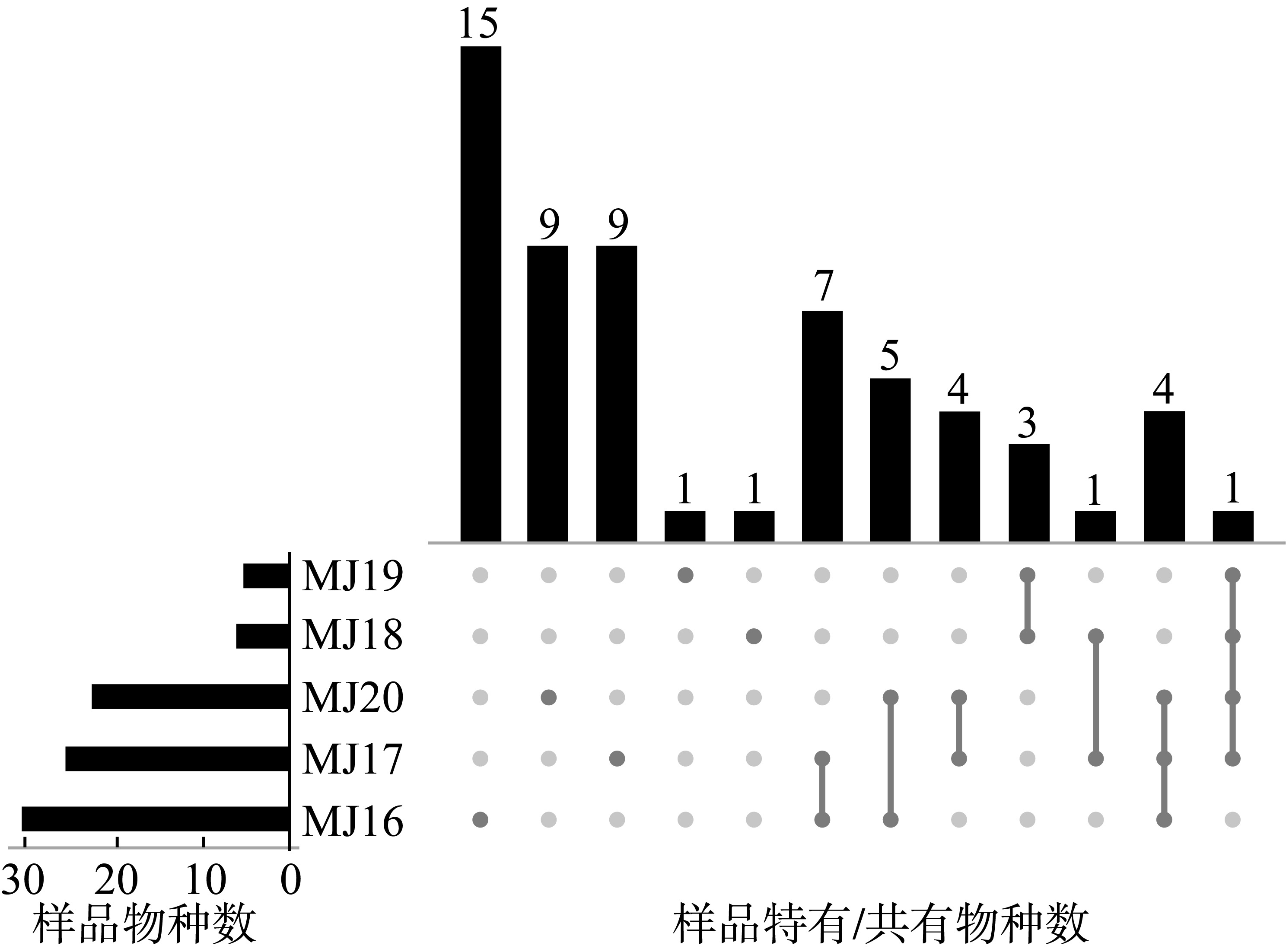
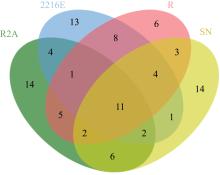
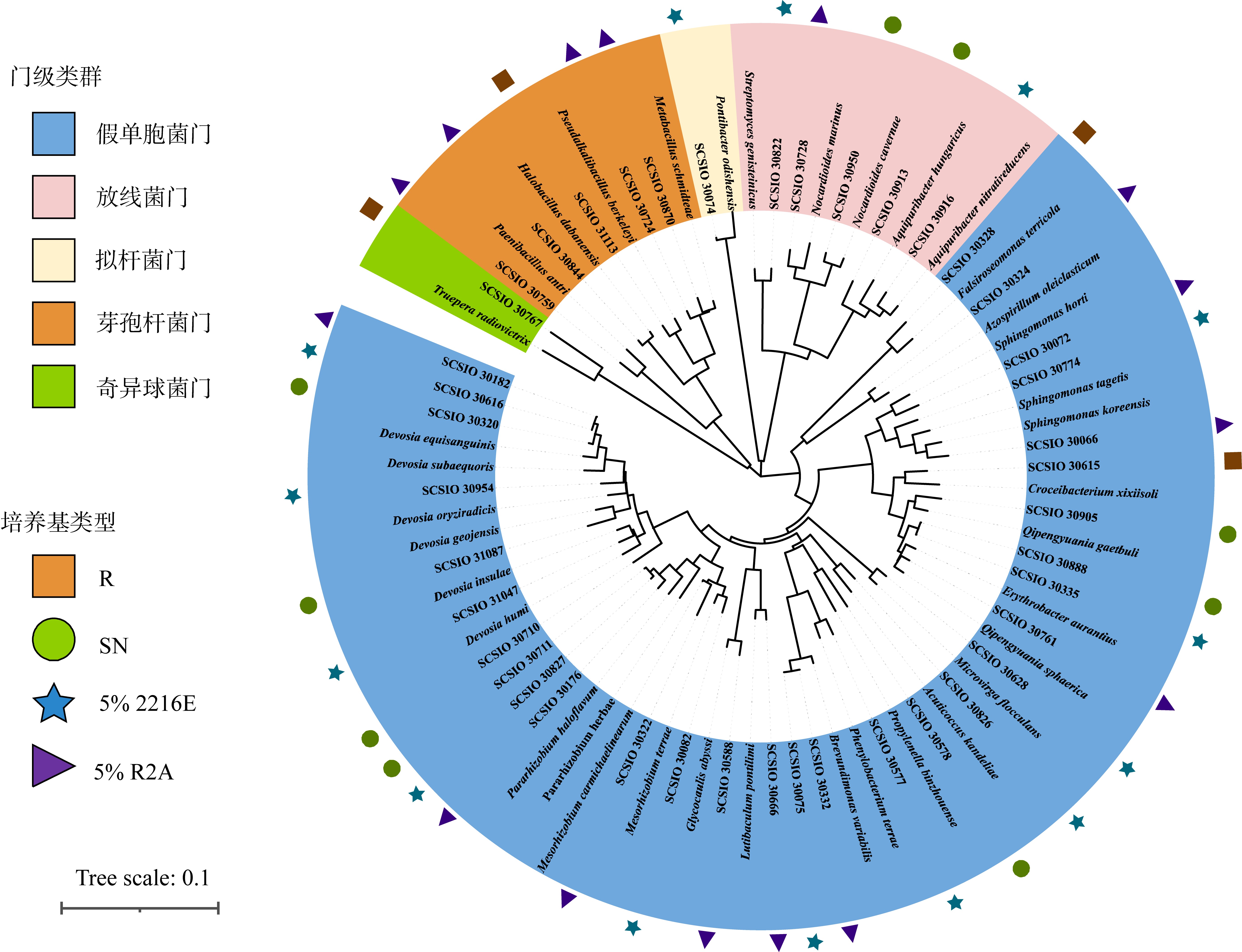
| [1] |
韩敏敏, 李蜜, 刘昕明, 等, 2020. Khai岛和Pathiu岛珊瑚礁沉积物细菌多样性及细菌粗提物延缓秀丽隐杆线虫衰老活性研究[J]. 热带海洋学报, 39(5): 19-29.
doi: 10.11978/2019126
|
|
HAN MINMIN, LI MI, LIU XIMING, et al, 2020. Studies on bacterial diversity in coral reef sediments in Khai Island and Pathiu Island and bacterial crude extract retards aging activity of Caenorhabditis elegans[J]. Journal of Tropical Oceanography, 39(5): 19-29 (in Chinese with English abstract).
|
| [2] |
何媛秋, 李存, 陈柔雯, 等, 2020. 不同培养条件对海洋沉积环境细菌的选择性分离[J]. 生物资源, 42(5): 540-548.
|
|
HE YUANQIU, LI CUN, CHEN ROUWEN, et al, 2020. Selective isolation of bacteria from marine sedimentary environment by different culture conditions[J]. Biotic Resources, 42(5): 540-548 (in Chinese with English abstract).
|
| [3] |
李存, 崔林青, 杨红强, 等, 2022. 三份南海岛礁珊瑚砂样品中可培养细菌多样性[J]. 热带海洋学报, 41(2): 149-158.
doi: 10.11978/2021050
|
|
LI CUN, CUI LINQING, YANG HONGQIANG, et al, 2022. Diversity of cultured bacteria isolated from three coral reef sediments in South China Sea[J]. Journal of Tropical Oceanography, 41(2): 149-158 (in Chinese with English abstract).
doi: 10.11978/2021050
|
| [4] |
刘津江, 王淼, 樊敏, 等, 2022. 产脲酶微生物的筛选和应用研究进展[J]. 生物技术, 32(1): 107-113, 119.
|
|
LIU JINJIANG, WANG MIAO, FAN MIN, et al, 2022. Research advances in application and screening of urease-producing microorganisms[J]. Biotechnology, 32(1): 107-113, 119 (in Chinese with English abstract).
|
| [5] |
柳鑫鹏, 臧淑英, 智刚, 等, 2022. 盐碱土耐盐碱细菌筛选及其植物促生能力研究[J]. 土壤通报, 53(3): 567-576.
|
|
LIU XINPENG, ZANG SHUYING, ZHI GANG, et al, 2022. Isolation for plant-growth promoting halotolerant bacteria from alkali-saline soil[J]. Chinese Journal of Soil Science, 53(3): 567-576 (in Chinese with English abstract).
|
| [6] |
邱路凡, 2023. 一株高效解钾菌解钾机理的探究及对土壤的改良[D]. 大庆: 东北石油大学.
|
|
QIU LUFAN, 2023. Study on the mechanism of potassium solubilization by a highly efficient potassium solubilizing bacterium and its improvement to soil[D]. Daqing: Northeast Petroleum University (in Chinese with English abstract).
|
| [7] |
王效昌, 马凯, 谢嘉慧, 等, 2023. 无机解磷菌对天鹅湖瀉湖沉积物内源磷释放的影响[J]. 农业资源与环境学报, 40(1): 76-85.
|
|
WANG XIAOCHANG, MA KAI, XIE JIAHUI, et al, 2023. Effects of inorganic phosphate-solubilizing bacteria on phosphorus release from sediments in Swan Lagoon[J]. Journal of Agricultural Resources and Environment, 40(1): 76-85 (in Chinese with English abstract).
|
| [8] |
朱红惠, 黄力, 李文均, 等, 2021. 未/难培养微生物: 风正扬帆再起航, 共辉煌[J]. 微生物学报, 61(4):ⅰ–ⅱ.
|
|
ZHU HONGHUI, HUANG LI, LI WENJUN, et al, 2021. Uncultivated microorganisms: it is time to refocus and reflourish to the special issue “uncultivated microorganisms”[J]. Acta Microbiologica Sinica, 61(4):ⅰ–ⅱ (in Chinese with English abstract).
|
| [9] |
ANGLY F E, HEATH C, MORGAN T C, et al, 2016. Marine microbial communities of the Great Barrier Reef lagoon are influenced by riverine floodwaters and seasonal weather events[J]. PeerJ, 4: e1511.
|
| [10] |
DINSDALE E A, PANTOS O, SMRIGA S, et al, 2008. Microbial ecology of four coral atolls in the northern line islands[J]. PLoS One, 3(2): e1584.
|
| [11] |
HUG L A, BAKER B J, ANANTHARAMAN K, et al, 2016. A new view of the tree of life[J]. Nature Microbiology, 1: 16048.
|
| [12] |
KRAMER J, ÖZKAYA Ö, KÜMMERLI R, 2020. Bacterial siderophores in community and host interactions[J]. Nature Reviews Microbiology, 18(3): 152-163.
doi: 10.1038/s41579-019-0284-4
pmid: 31748738
|
| [13] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096
pmid: 29722887
|
| [14] |
LETUNIC I, BORK P, 2019. Interactive tree of life (iTOL) v4: recent updates and new developments[J]. Nucleic Acids Research, 47(W1): W256-W259.
doi: 10.1093/nar/gkz239
|
| [15] |
LIU LIRUI, HUANG WENCONG, PAN JIE, et al, 2023. Isolation and genomics of Futiania mangrovii gen. nov., sp. nov., a rare and metabolically versatile member in the class Alphaproteobacteria[J]. Microbiology Spectrum, 11(1): e0411022.
|
| [16] |
MOLITOR R, BOLLINGER A, KUBICKI S, et al, 2020. Agar plate-based screening methods for the identification of polyester hydrolysis by Pseudomonas species[J]. Microbial Biotechnology, 13(1): 274-284.
|
| [17] |
OUILLON S, DOUILLET P, LEFEBVRE J P, et al, 2010. Circulation and suspended sediment transport in a coral reef lagoon: the south-west lagoon of New Caledonia[J]. Marine Pollution Bulletin, 61(7-12): 269-296.
doi: 10.1016/j.marpolbul.2010.06.023
pmid: 20637477
|
| [18] |
PEIXOTO R S, ROSADO P M, DE ASSIS LEITE D C, et al, 2017. Beneficial microorganisms for corals (BMC): proposed mechanisms for coral health and resilience[J]. Frontiers in Microbiology, 8: 341.
doi: 10.3389/fmicb.2017.00341
pmid: 28326066
|
| [19] |
QIN ZHENJUN, YU KEFU, CHEN SHUCHANG, et al, 2021. Microbiome of juvenile corals in the outer reef slope and lagoon of the South China Sea: insight into coral acclimatization to extreme thermal environments[J]. Environmental Microbiology, 23(8): 4389-4404.
doi: 10.1111/1462-2920.15624
pmid: 34110067
|
| [20] |
SAITOU N, NEI M, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees[J]. Molecular Biology and Evolution, 4(4): 406-425.
doi: 10.1093/oxfordjournals.molbev.a040454
pmid: 3447015
|
| [21] |
WALSH P S, METZGER D A, HIGUCHI R, 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material[J]. Biotechniques, 10(4): 506-513.
pmid: 1867860
|
| [22] |
YANG DIHE, TANG LU, CUI YING, et al, 2022. Saline-alkali stress reduces soil bacterial community diversity and soil enzyme activities[J]. Ecotoxicology, 31(9): 1356-1368.
doi: 10.1007/s10646-022-02595-7
pmid: 36208367
|
| [23] |
YARZA P, YILMAZ P, PRUESSE E, et al, 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences[J]. Nature Reviews Microbiology, 12(9): 635-645.
doi: 10.1038/nrmicro3330
pmid: 25118885
|
| [24] |
YIN WEN, WANG YITING, LIU LU, et al, 2019. Biofilms: the microbial "protective clothing" in extreme environments[J]. International Journal of Molecular Sciences, 20(14): 3423.
|
| [25] |
YOON S H, HA S M, KWON S, et al, 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies[J]. International Journal of Systematic and Evolutionary Microbiology, 67(5): 1613-1617.
|
 ), 简丽丽1,2,3, 石松标1,2,3, 田新朋1,2(
), 简丽丽1,2,3, 石松标1,2,3, 田新朋1,2( )
)
 ), JIAN Lili1,2,3, SHI Songbiao1,2,3, TIAN Xinpeng1,2(
), JIAN Lili1,2,3, SHI Songbiao1,2,3, TIAN Xinpeng1,2( )
)