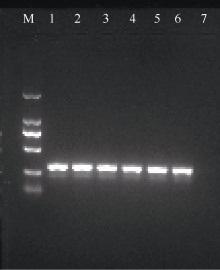
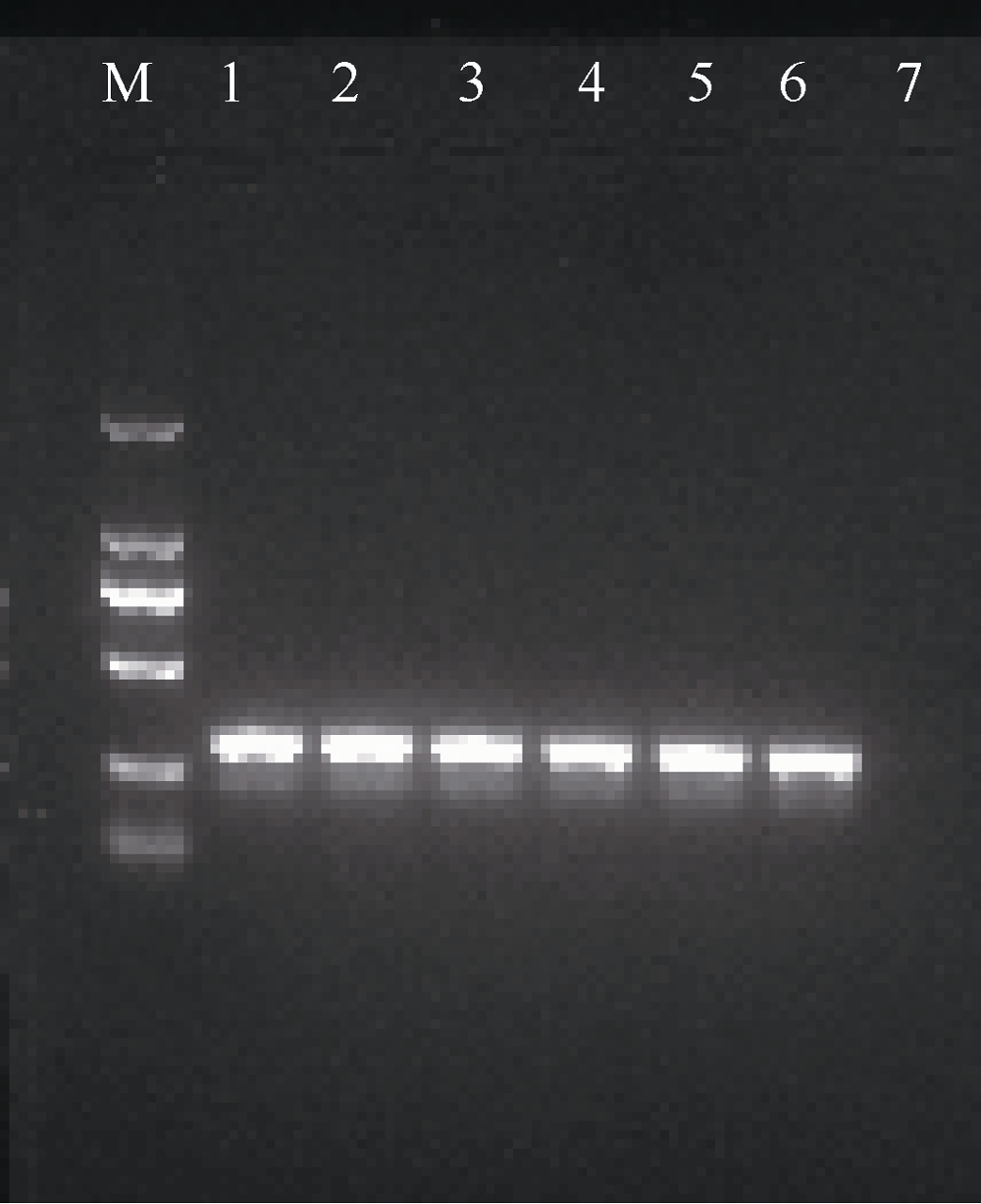
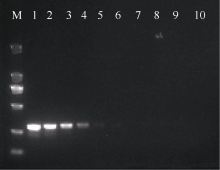
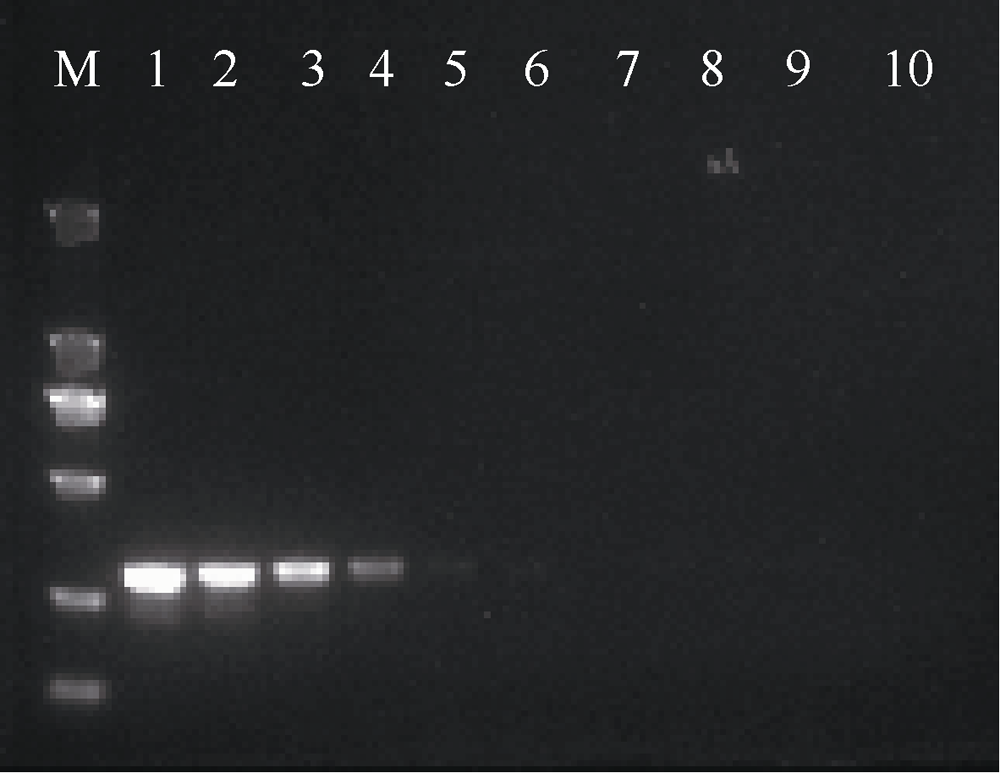
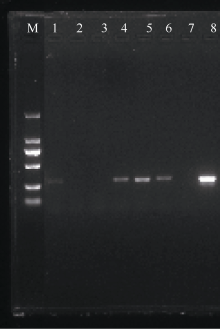
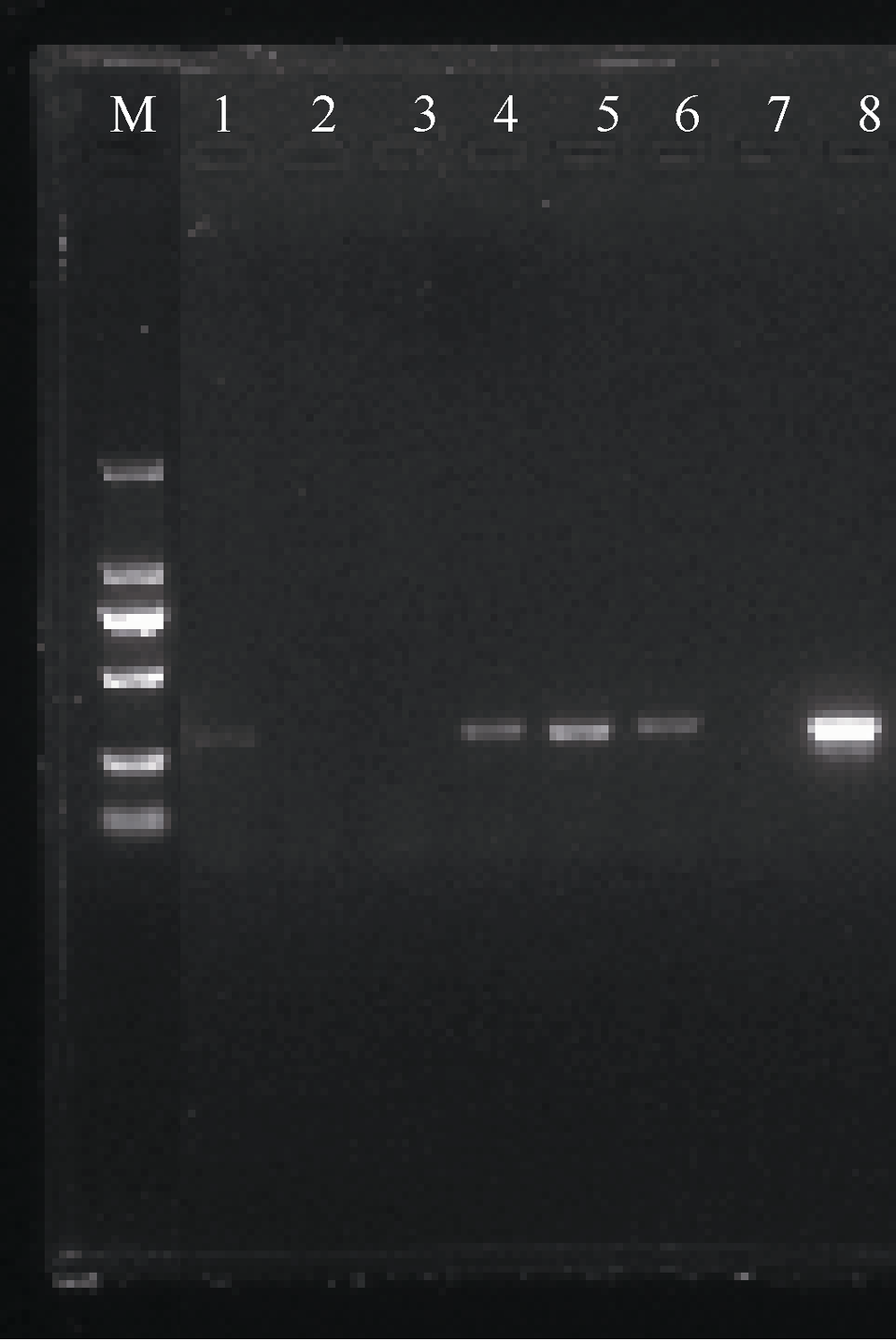
| [1] |
李元超, 吴钟解, 梁计林, 等, 2019. 近15年西沙群岛长棘海星暴发周期及暴发原因分析[J]. 科学通报, 64(33): 3478-3484.
|
|
LI YUANCHAO, WU ZHOGNJIE, LIANG JILIN, et al, 2019. Analysis on the outbreak period and cause of Acanthaster planci in Xisha Islands in recent 15 years[J]. Chinese Science Bulletin, 64(33): 3478-3484. (in Chinese with English abstract)
|
| [2] |
位正鹏, 孔晓瑜, 吴相云, 等, 2009. 基于COI序列的翡翠股贻贝Perna viridis线粒体遗传特性分析及其近缘种间的系统关系探讨[J]. 热带海洋学报, 28(6): 72-78.
doi: 10.11978/j.issn.1009-5470.2009.06.072
|
|
WEI ZHENGPENG, KONG XIAOYU, WU XIANGYUN, et al, 2009. Genetic analysis of the marine mussel Perna viridis (Bivalvia: Mytilidae) based on mitochondrial COI sequence and its phylogenetic relationship with other congeneric species[J]. Journal of Tropical Oceanography, 28(6): 72-78. (in Chinese with English abstract)
|
| [3] |
吴钟解, 王道儒, 涂志刚, 等, 2011. 西沙生态监控区造礁石珊瑚退化原因分析[J]. 海洋学报, 33(4): 140-146.
|
|
WU ZHOGNJIE, WANG DAORU, TU ZHIGANG, et al, 2011. The analysis on the reason of hermatypic coral degradation in Xisha[J]. Acta Oceanologica Sinica, 33(4): 140-146. (in Chinese with English abstract)
|
| [4] |
BAIRD A H, PRATCHETT M S, HOEY A S, et al, 2013. Acanthaster planci is a major cause of coral mortality in Indonesia[J]. Coral Reefs, 32(3): 803-812.
doi: 10.1007/s00338-013-1025-1
|
| [5] |
BUCKLIN A, LINDEQUE P K, RODRIGUEZ-EZPELETA N, et al, 2016. Metabarcoding of marine zooplankton: Prospects, progress and pitfalls[J]. Journal of Plankton Research, 38(3): 393-400.
doi: 10.1093/plankt/fbw023
|
| [6] |
DE'ATH G, FABRICIUS K E, SWEATMAN H, et al, 2012. The 27-year decline of coral cover on the Great Barrier Reef and its causes[J]. Proceedings of the National Academy of Sciences of the United States of America, 109(44): 17995-17999.
doi: 10.1073/pnas.1208909109
pmid: 23027961
|
| [7] |
DIGHT I J, JAMES M K, BODE L, 1990. Modelling the larval dispersal of Acanthaster planci II. Patterns of reef connectivity[J]. Coral Reefs, 9(3): 125-134.
doi: 10.1007/BF00258224
|
| [8] |
DOYLE J R, MCKINNON A D, UTHICKE S, 2017. Quantifying larvae of the coralivorous seastar Acanthaster cf. solaris on the Great Barrier Reef using qPCR[J]. Marine Biology, 164(8): 176.
doi: 10.1007/s00227-017-3206-x
|
| [9] |
GÉRARD K, ROBY C, CHEVALIER N, et al, 2008. Assessment of three mitochondrial loci variability for the crown-of-thorns starfish: A first insight into Acanthaster phylogeography[J]. Comptes Rendus Biologies, 331(2): 137-143.
doi: 10.1016/j.crvi.2007.11.005
|
| [10] |
HEBERT P D N, RATNASINGHAM S, DE WAARD J R, 2003. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species[J]. Proceedings of the Royal Society B-Biological Sciences, 270(S1): S96-S99.
|
| [11] |
HUGHES T P, ANDERSON K D, CONNOLLY S R, et al, 2018. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene[J]. Coral Reefs, 359(6371): 80-83.
|
| [12] |
HUGHES T P, BARNES M L, BELLWOOD D R, et al, 2017. Coral reefs in the Anthropocene[J]. Nature, 546(7656): 82-90.
doi: 10.1038/nature22901
|
| [13] |
KAYAL M, VERCELLONI J, LISON DE LOMA T, et al, 2012. Predator crown-of-thorns starfish (Acanthaster planci) outbreak, mass mortality of corals, and cascading effects on reef fish and benthic communities[J]. PLoS One, 7(10): e47363.
doi: 10.1371/journal.pone.0047363
|
| [14] |
NAKAMURA M, OKAJI K, HIGA Y, et al, 2014. Spatial and temporal population dynamics of the crown-of-thorns starfish, Acanthaster planci, over a 24-year period along the central west coast of Okinawa Island, Japan[J]. Marine Biology, 161(11): 2521-2530.
doi: 10.1007/s00227-014-2524-5
|
| [15] |
PRATCHETT M S, CABALLES C F, RIVER-POSADA J A, et al, 2014. Limits to understanding and managing outbreaks of Crown- of- Thorns Starfish (Acanthaster spp.)[M]// HUGHESR N, HUGHESD J, SMITHI P. Oceanography and Marine Biology: An Annual Review, Volume 52. Boca Raton: CRC Press: 68.
|
| [16] |
PRATCHETT M S, CABALLES C F, WILMES J C, et al, 2017. Thirty years of research on crown-of-thorns starfish (1986- 2016): Scientific advances and emerging opportunities[J]. Diversity, 9(4): 41.
doi: 10.3390/d9040041
|
| [17] |
SAMEOTO J A, METAXAS A, 2008. Interactive effects of haloclines and food patches on the vertical distribution of 3 species of temperate invertebrate larvae[J]. Journal of Experimental Marine Biology and Ecology, 367(2): 131-141.
doi: 10.1016/j.jembe.2008.09.003
|
| [18] |
SAPONARI L, MONTANO S, SEVESO D, et al, 2015. The occurrence of an Acanthaster planci outbreak in Ari Atoll, Maldives[J]. Marine Biodiversity, 45(4): 599-600.
doi: 10.1007/s12526-014-0276-6
|
| [19] |
SUZUKI G, YASUDA N, IKEHARA K, et al, 2016. Detection of a high-density brachiolaria-stage larval population of crown-of-thorns sea star (Acanthaster planci) in Sekisei Lagoon (Okinawa, Japan)[J]. Diversity, 8(2): 9.
doi: 10.3390/d8020009
|
| [20] |
TIAN Y S, YU Z L, LUO P, et al, 2017. Artificial breeding and culture of the crown-of-thorns starfish Acanthaster planci (Linnaeus, 1758) larvae in China[J]. Invertebrate Reproduction & Development, 61(3): 157-163.
|
| [21] |
UTHICKE S, DOYLE J, DUGGAN S, et al, 2015. Outbreak of coral-eating Crown-of-Thorns creates continuous cloud of larvae over 320 km of the Great Barrier Reef[J]. Scientific Reports, 5: 16885.
doi: 10.1038/srep16885
|
| [22] |
WARD R D, HOLMES B H, O'HARA T D, 2008. DNA barcoding discriminates echinoderm species[J]. Molecular Ecology Resources, 8(6): 1202-1211.
doi: 10.1111/j.1755-0998.2008.02332.x
pmid: 21586007
|
| [23] |
WOLANSKI E, HAMNER W M, 1988. Topographically controlled fronts in the ocean and their biological influence[J]. Science, 241(4862): 177-181.
pmid: 17841048
|
| [24] |
WOLFE K, GRABA-LANDRY A, DWORJANYN S A, et al, 2015. Larval phenotypic plasticity in the boom-and-bust crown- of-thorns seastar, Acanthaster planci[J]. Marine Ecology Progress Series, 539: 179-189.
doi: 10.3354/meps11495
|
| [25] |
WOOLDRIDGE S A, BRODIE J E, 2015. Environmental triggers for primary outbreaks of crown-of-thorns starfish on the Great Barrier Reef, Australia[J]. Marine Pollution Bulletin, 101(2): 805-815.
doi: 10.1016/j.marpolbul.2015.08.049
pmid: 26460182
|
| [26] |
YE J, COULOURIS G, ZARETSKAYA I, et al, 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction[J]. BMC Bioinformatics, 13(1): 134.
doi: 10.1186/1471-2105-13-134
|
 ), YANG Litong2, LIU Bing1,3, ZHENG Fanyu1, LUO Peng1, CHEN Chang1,4(
), YANG Litong2, LIU Bing1,3, ZHENG Fanyu1, LUO Peng1, CHEN Chang1,4( )
)