热带海洋学报 ›› 2022, Vol. 41 ›› Issue (6): 44-55.doi: 10.11978/2022061CSTR: 32234.14.2022061
低温胁迫下红榄李(Lumnitzera littorea)DEAD-box RNA解旋酶基因的表达分析
郝露露1,2( ), 柯明思2, 朱奕秀2, 许燕敏2, 张颖2,3, 郑春芳1(
), 柯明思2, 朱奕秀2, 许燕敏2, 张颖2,3, 郑春芳1( )
)
- 1.温州大学生命与环境科学学院, 浙江 温州 325035
2.岭南师范学院生命科学与技术学院, 广东 湛江 524048
3.海南省林业科学研究(海南省红树林研究院), 海南 海口 571129
-
收稿日期:2022-03-29修回日期:2022-05-23出版日期:2022-11-10发布日期:2022-05-31 -
通讯作者:郑春芳 -
作者简介:郝露露, 女(1996—), 山西省临汾市人, 硕士研究生, 从事红树植物的抗逆研究。email: 1973497638@qq.com -
基金资助:海南省林业科学研究院(海南省红树林研究院)基础性科研工作(KYYS-2021-04);海南省科研院所技术创新专项(KYYS-2021-13);海南省科研院所技术创新专项(KYYS-2021-22);海南省科研院所技术创新专项基础性科研工作项目(jcxk202003);国家自然科学基金(32071503)
Expression of DEAD-box RNA helicase enzyme genes in Lumnitzera littorea under low temperature stress
HAO Lulu1,2( ), KE Mingsi2, ZHU Yixiu2, XU Yanmin2, ZHANG Ying2,3, ZHENG Chunfang1(
), KE Mingsi2, ZHU Yixiu2, XU Yanmin2, ZHANG Ying2,3, ZHENG Chunfang1( )
)
- 1. College of Life and Environmental Science, Wenzhou University, Wenzhou 325035, China
2. School of Life Sciences and Technology, Lingnan Normal University, Zhanjiang 524048, China
3. Forestry Science Research of Hainan Province (Hainan Mangrove Research Institute), Haikou 571129, China
-
Received:2022-03-29Revised:2022-05-23Online:2022-11-10Published:2022-05-31 -
Contact:ZHENG Chunfang -
Supported by:Basic scientific research work of Hainan Forestry Research Institute (Hainan Mangrove Research Institute)(KYYS-2021-04);Technological Innovation Special project of Hainan Scientific Research Institute(KYYS-2021-13);Technological Innovation Special project of Hainan Scientific Research Institute(KYYS-2021-22);Special Basic Research Work Project for Technological Innovation in Hainan Research Institutes(jcxk202003);National Natural Science Foundation of China(32071503)
摘要:
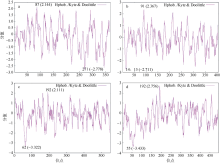
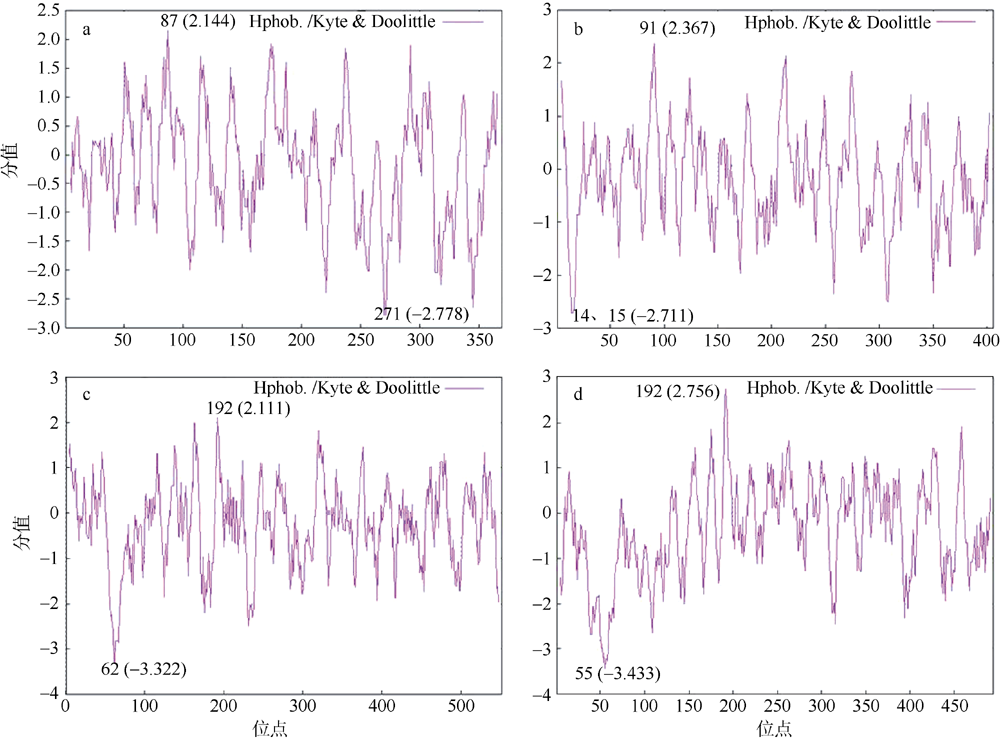
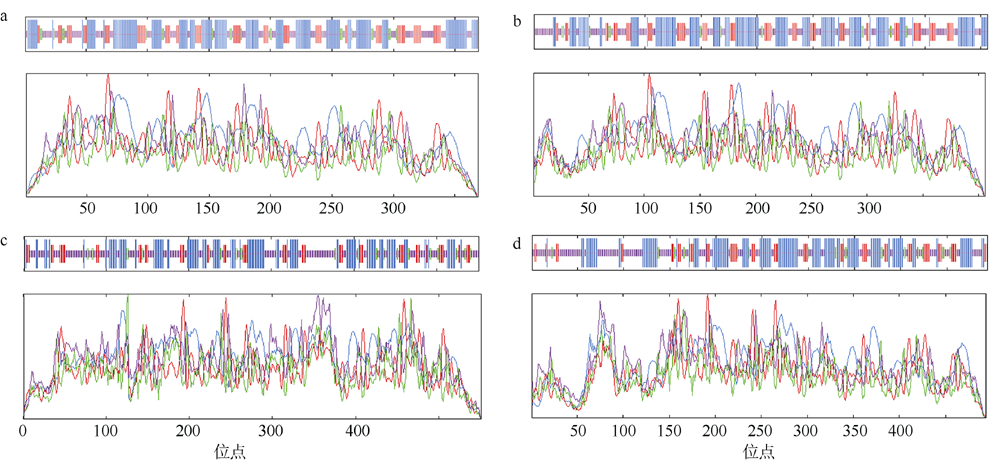
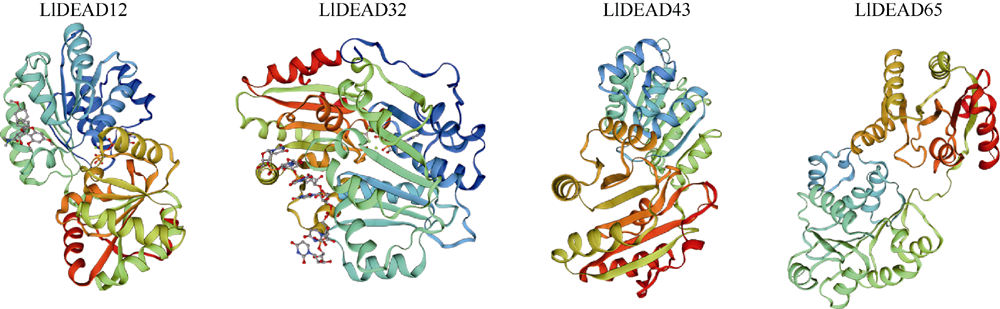
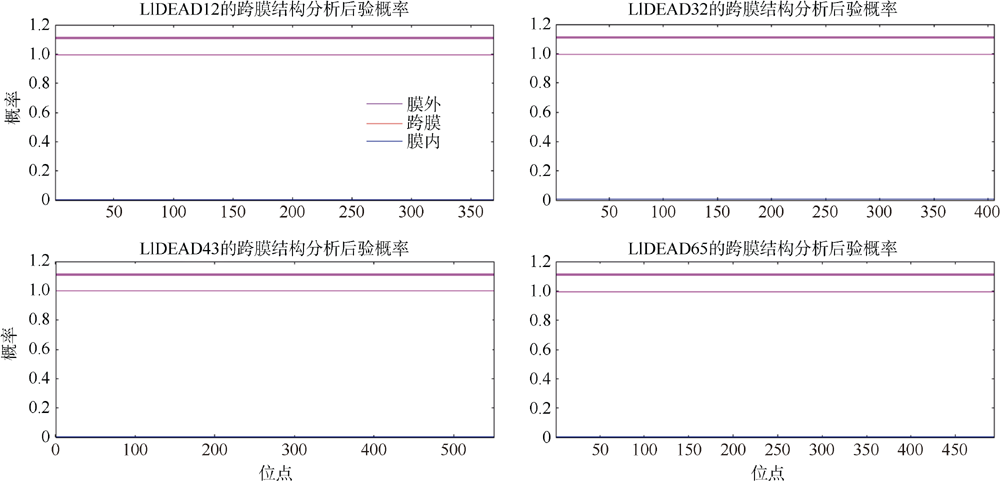
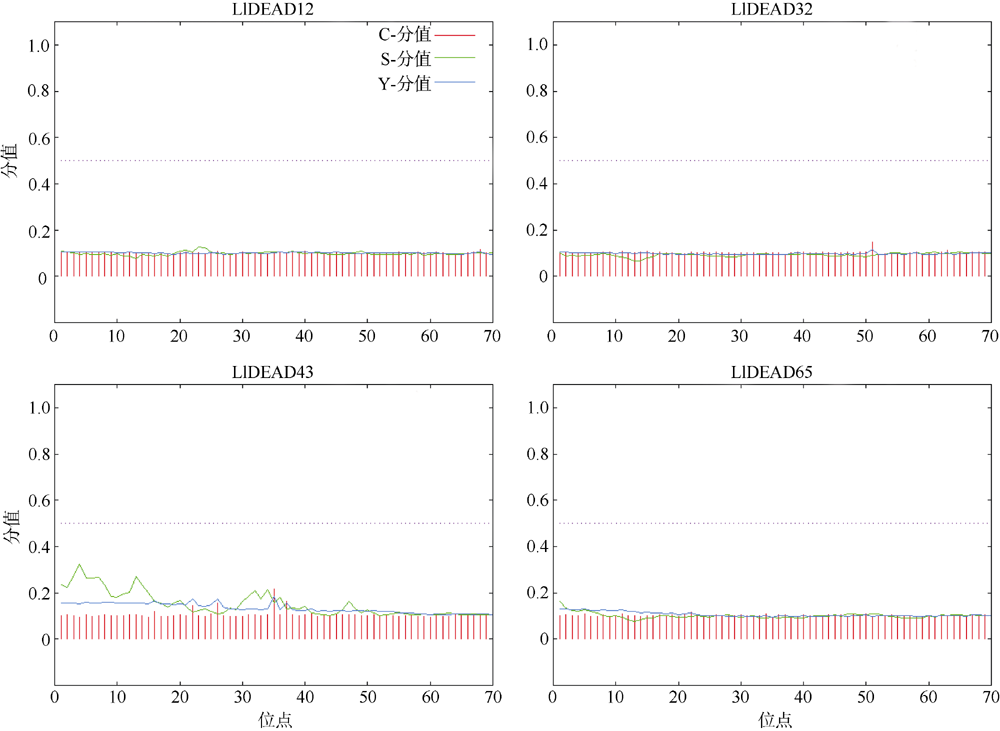
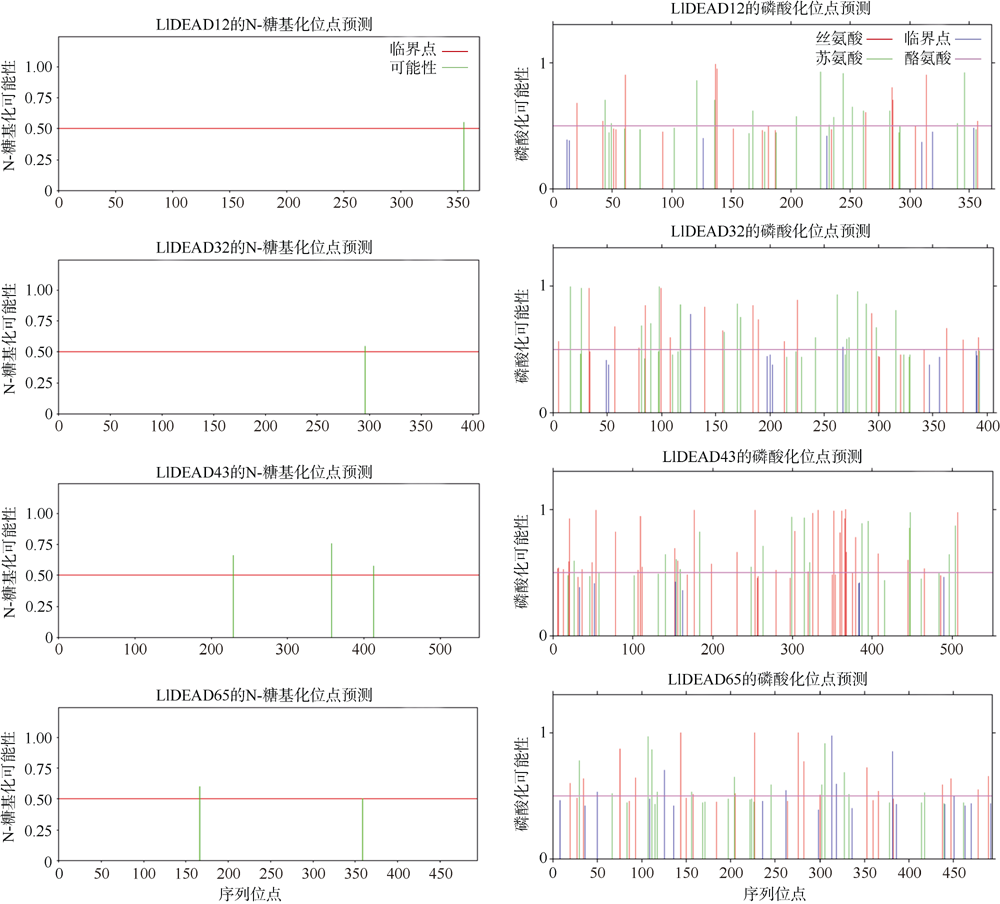
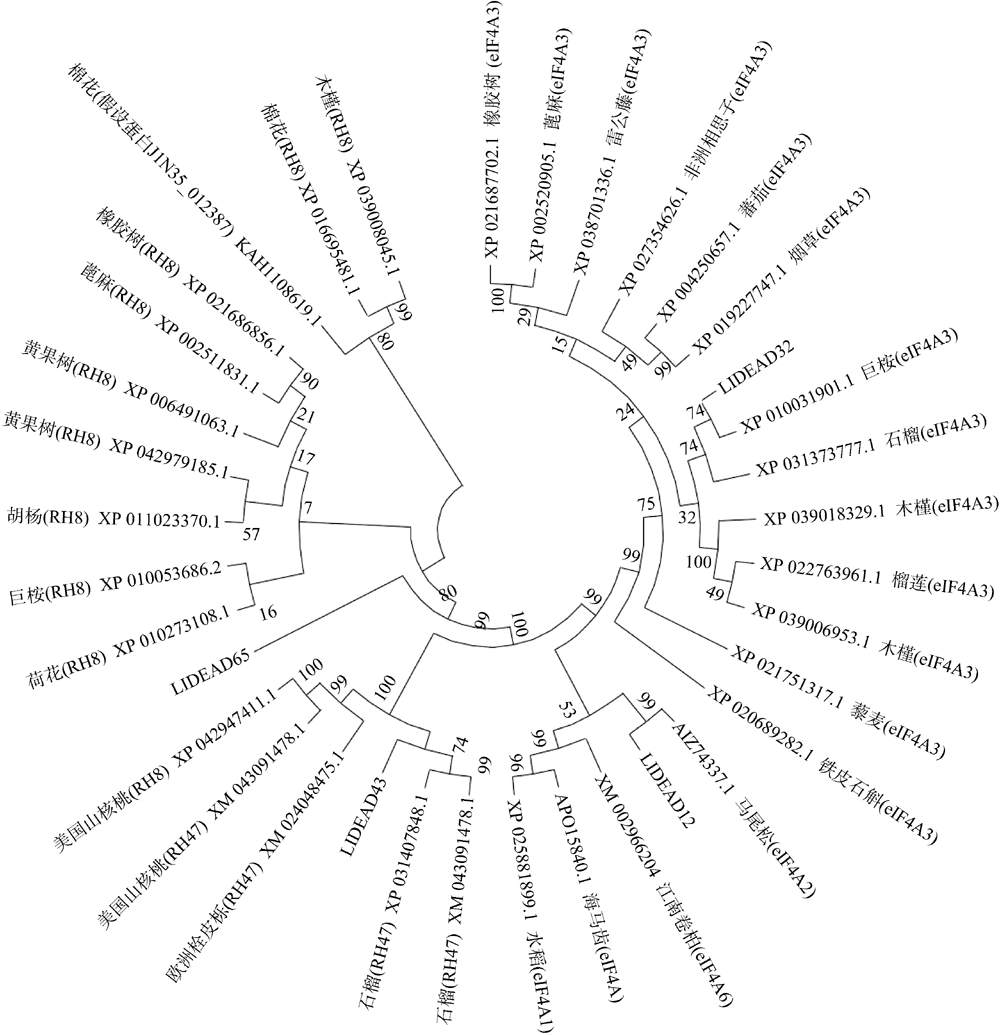
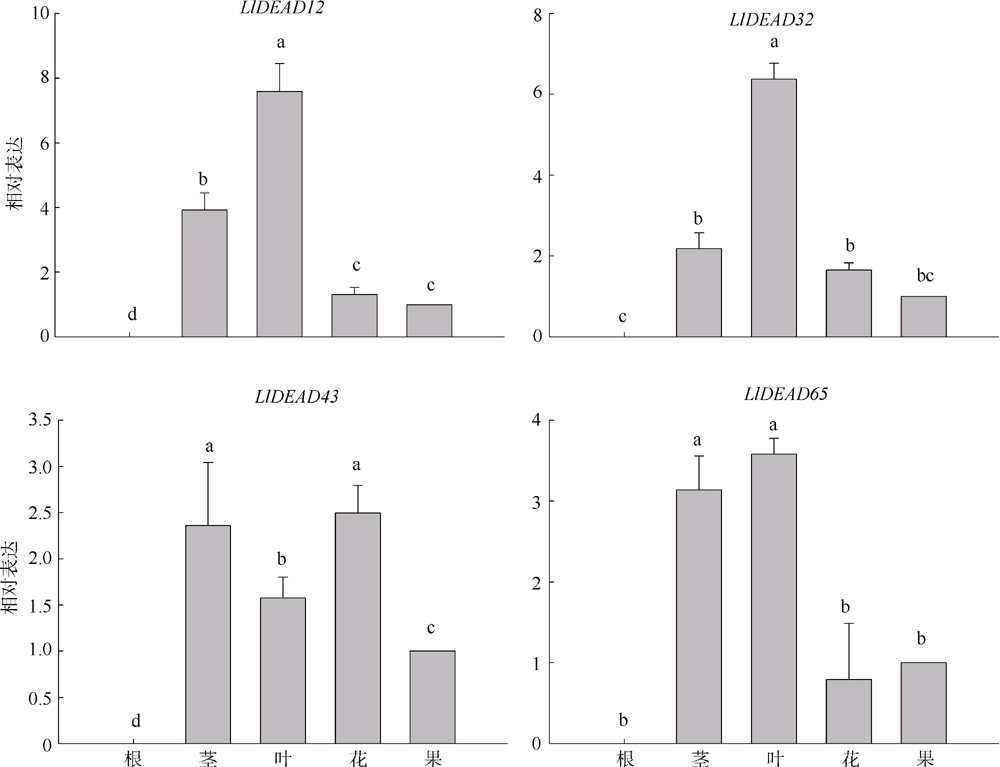
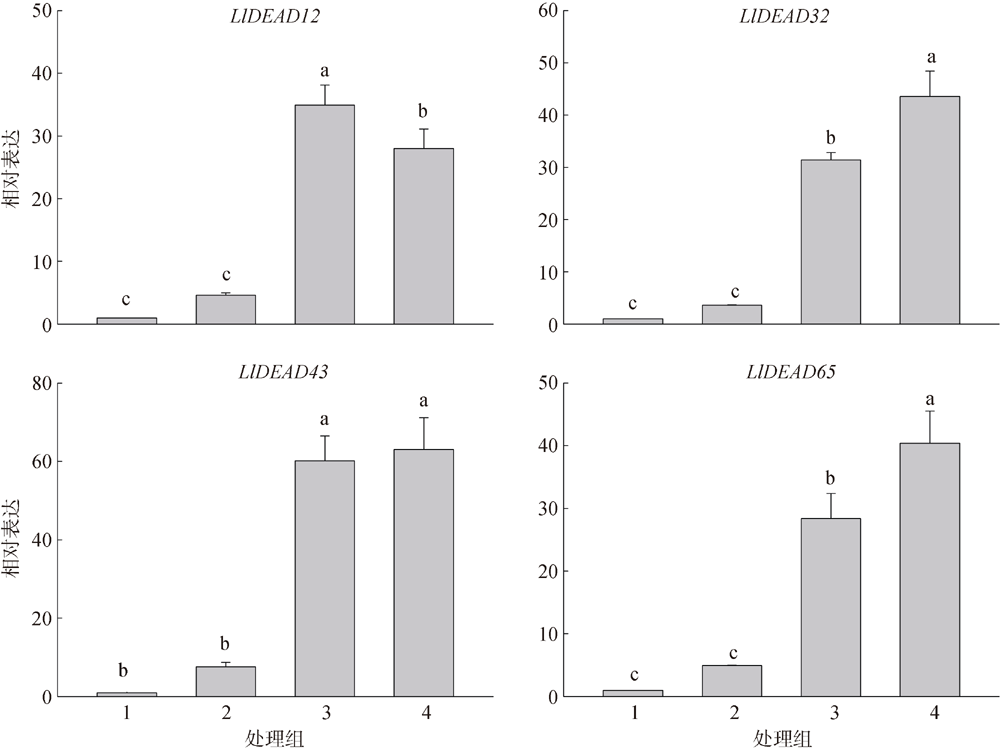
转录组分析发现DEAD-box RNA解旋酶家族参与红榄李(Lumnitzera littorea)对低温胁迫的响应。本文对4个低温显著差异表达基因LlDEAD12, LlDEAD32, LlDEAD43和LlDEAD65的生物信息学特性、组织特异性以及在不同温度处理下的红榄李幼苗中的差异表达进行研究。结果表明, LlDEAD12、LlDEAD32、LlDEAD43和LlDEAD65均属于疏水性蛋白, 二级结构均由α-螺旋、β-转角、延伸链和无规则卷曲组成, 具有多个糖基化位点和磷酸化位点, 且不含有跨膜结构域和信号肽。亚细胞定位分析表明, LlDEAD12和LlDEAD32分别定位于细胞质和细胞核, 而LlDEAD43和LlDEAD65则定位于线粒体上。通过蛋白质氨基酸序列的比对分析发现LlDEAD12、LlDEAD32和LlDEAD43分别与马尾松(Pinus massoniana )、巨桉 (Eucalyptus grandis)和石榴 (Punica granatum)的DEAD-box RNA解旋酶有较近的亲缘关系。荧光定量PCR技术分析发现, LlDEAD12和LlDEAD32在叶中高表达, LlDEAD43和LlDEAD65在茎和花中高表达, 但这4个基因在根中都不表达, 说明这4个基因对红榄李生长发育的调控主要集中在叶、茎和花等器官。低温胁迫对红榄李幼苗这4个基因的表达均有显著的抑制, 说明它们参与了红榄李在低温环境下的分子响应, 其中LlDEAD12和LlDEAD32可能参与了叶绿体的发育, LlDEAD43和LlDEAD65可能参与到了维持线粒体功能的稳定。以上结果可为红榄李抗冷性苗木的培育提供科学依据。
中图分类号:
- Q943.2
引用本文
郝露露, 柯明思, 朱奕秀, 许燕敏, 张颖, 郑春芳. 低温胁迫下红榄李(Lumnitzera littorea)DEAD-box RNA解旋酶基因的表达分析[J]. 热带海洋学报, 2022, 41(6): 44-55.
HAO Lulu, KE Mingsi, ZHU Yixiu, XU Yanmin, ZHANG Ying, ZHENG Chunfang. Expression of DEAD-box RNA helicase enzyme genes in Lumnitzera littorea under low temperature stress[J]. Journal of Tropical Oceanography, 2022, 41(6): 44-55.
表1
在线分析的网站及网址"
| 网站 | 功能 | 网址链接 | 参数设置 |
|---|---|---|---|
| ExPASy ProtParam | 理化性质分析 | https://web.expasy.org/protparam/ | 默认参数 |
| PSORT | 亚细胞定位分析 | https://www.genscript.com/psort.html?src=leftbar | 默认参数 |
| ProtScale | 亲水性分析 | https://web.expasy.org/cgi-bin/protscale/protscale.pl | Hphob./Kyte & Doolittle, Window size: 9; Weight variation model:linear |
| SOPMA | 二级结构分析 | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html | Number of conformational states: 4; Similarity threshold: 8; Window width: 17 |
| TMHMM | 跨膜结构分析 | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 默认参数 |
| SWISS-MODEL | 三级结构分析 | https://swissmodel.expasy.org/interactive | 默认参数 |
| SignalP4.1 | 信号肽分析 | https://services.healthtech.dtu.dk/service.php?SignalP-4.1 | Organism group: Eukaryotes; D-cutoff values: Default; Output format: Standard; Method: Input sequences may include TM regions |
| NetPhos3.1 | 磷酸化位点分析 | https://services.healthtech.dtu.dk/service.php?NetPhos-3.1 | Residues to predict: all three; |
| NetNGlyc1.0 | 糖基化位点分析 | https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0 | 默认设置 |
表2
qPCR引物序列"
| 引物 | 引物序列(5′-3′) | 引物长度/bp |
|---|---|---|
| AthU6-F | ACATCCGATAAAATTGGAACGA | 168 |
| AthU6-R | TTTTTTTGGACCATTTCTCGAT | |
| LlDEAD12-F | GGCTATGACTACCGCAATCTC | 237 |
| LlDEAD12-R | GCCCTTTCAACACTATGCTTCT | |
| LlDEAD32-F | CTGTTGTGCTCTGCCCCA | 204 |
| LlDEAD32-R | CCATGTTCCCATCTTCAATATG | |
| LlDEAD43-F | ATCCTTCCATAGGCGTTCA | 105 |
| LlDEAD43-R | TTCCAGGGGTAGCCACAA | |
| LlDEAD65-F | CGGGCAGATTGGTGGATT | 165 |
| LlDEAD65-R | TGGTCTGTCTTTCACCTCGG |
表4
红榄李DEAD-box蛋白理化性质分析及亚细胞定位分析"
| 理化指标 | 蛋白质 | |||
|---|---|---|---|---|
| LlDEAD12 | LlDEAD32 | LlDEAD43 | LlDEAD65 | |
| 氨基酸的数量 | 1546 | 2619 | 2144 | 3302 |
| 分子量/Da | 129345.23 | 220714.18 | 176455.64 | 276389.07 |
| 等电点 | 4.94 | 4.82 | 4.94 | 4.82 |
| 稳定指数 | 56.86 | 51.35 | 38.34 | 47.32 |
| 脂肪指数 | 27.04 | 31.54 | 29.24 | 31.8 |
| 亲水性总平均值 | 0.915 | 0.882 | 0.711 | 0.864 |
| 原子总数 | 16283 | 28285 | 22738 | 35465 |
| 亚细胞定位 | 细胞质 | 细胞核 | 线粒体 | 线粒体 |
| [1] | 蔡敬, 孟小庆, 董婷婷, 等, 2017. DEAD-box解旋酶在植物非生物胁迫响应中的功能研究进展[J]. 生命科学, 29(5): 427-433. |
| CAI JING, MENG XIAOQING, DONG TINGTING, et al, 2017. Progress of plant DEAD-box helicase in response to abiotic stress[J]. Chinese Bulletin of Life Sciences, 29(5): 427-433. (in Chinese with English abstract) | |
| [2] | 陈丹, 彭雄波, 2016. DEAD-box基因家族在拟南芥生长发育中的作用[J]. 植物科学学报, 34(6): 941-948. |
| CHEN DAN, PENG XIONGBO, 2016. DEAD-box: function on growth and development in Arabidopsis thaliana[J]. Plant Science Journal, 34(6): 941-948. (in Chinese with English abstract) | |
| [3] | 孔荣荣, 2019. 水稻DEAD-box RNA解旋酶蛋白基因TCD33功能研究[D]. 上海: 上海师范大学. |
| KONG RONGRONG, 2019. Functional research of a DEAD-box RNA helicase gene TCD33 in rice (Oryza sativa L.)[D]. Shanghai: Shanghai Normal University. (in Chinese with English abstract) | |
| [4] | 铁原毓, 田洁, 2021. 大蒜蔗糖转化酶基因AsINV的克隆及其响应低温和干旱胁迫的表达分析[J]. 植物生理学报, 57(12): 2258-2270. |
| TIE YUANYU, TIAN JIE, 2021. Cloning of Allium sativum invertase gene AsINV and its expression analysis in response to low temperature and drought stress[J]. Plant Physiology Journal, 57(12): 2258-2270. (in Chinese with English abstract) | |
| [5] | 张颖, 陈光程, 钟才荣, 2021. 中国濒危红树植物研究与恢复现状[J]. 应用海洋学学报, 40(1): 142-153. |
| ZHANG YING, CHEN GUANGCHENG, ZHONG CAIRONG, 2021. Research on endangered mangrove species and recovery status in China[J]. Journal of Applied Oceanography, 40(1): 142-153.. (in Chinese with English abstract) | |
| [6] | 张颖, 钟才荣, 杨勇, 等, 2018. 濒危红树植物红榄李种质资源挽救[J]. 分子植物育种, 16(12): 4112-4118. |
| ZHANG YING, ZHONG CAIRONG, YANG YONG, et al, 2018. Rescue of germplasm resources of endangered mangrove plant Lumnitzera littorea[J]. Molecular Plant Breeding, 16(12): 4112-4118. (in Chinese with English abstract) | |
| [7] | 钟才荣, 李诗川, 管伟, 等, 2011. 中国3种濒危红树植物的分布现状[J]. 生态科学, 30(4): 431-435. |
| ZHONG CAIRONG, LI SHICHUAN, GUAN WEI, et al, 2011. Current distributions of three endangered mangrove species in China[J]. Ecological Science, 30(4): 431-435. (in Chinese with English abstract) | |
| [8] |
ANANTHARAMAN V, KOONIN E V, ARAVIND L, 2002. Comparative genomics and evolution of proteins involved in RNA metabolism[J]. Nucleic Acids Research, 30(7): 1427-1464.
pmid: 11917006 |
| [9] |
ANDERSON P, KEDERSHA N, 2006. RNA granules[J]. Journal of Cell Biology, 172(6): 803-808.
pmid: 16520386 |
| [10] |
ASAKURA Y, GALARNEAU E, WATKINS K P, et al, 2012. Chloroplast RH3 DEAD box RNA helicases in Maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis[J]. Plant Physiology, 159(3): 961-974.
doi: 10.1104/pp.112.197525 pmid: 22576849 |
| [11] |
BRAUD C, ZHENG WENGUANG, XIAO WENYAN, 2012. LONO1 encoding a nucleoporin is required for embryogenesis and seed viability in Arabidopsis[J]. Plant Physiology, 160(2): 823-836.
doi: 10.1104/pp.112.202192 |
| [12] |
CHEN PENGYUN, JIAN HONGLIANG, WEI FEI, et al, 2021. Phylogenetic analysis of the membrane attack complex/ perforin domain-containing proteins in Gossypium and the role of GhMACPF26 in cotton under cold stress[J]. Frontiers in Plant Science, 12: 684227.
doi: 10.3389/fpls.2021.684227 |
| [13] |
CHI WEI, HE BAOYE, MAO JUAN, et al, 2012. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts[J]. Plant Physiology, 158(2): 693-707.
doi: 10.1104/pp.111.186775 pmid: 22170977 |
| [14] |
CORDIN O, BANROQUES J, TANNER N K, et al, 2006. The DEAD-box protein family of RNA helicases[J]. Gene, 367: 17-37.
pmid: 16337753 |
| [15] |
FAIRMAN-WILLIAMS M E, GUENTHER U P, JANKOWSKY E, 2010. SF1 and SF2 helicases: family matters[J]. Current Opinion in Structural Biology, 20(3): 313-324.
doi: 10.1016/j.sbi.2010.03.011 |
| [16] |
GE SHASHA, DUO LAN, WANG JUNQI, et al, 2021. A unique understanding of traditional medicine of pomegranate, Punica granatum L. and its current research status[J]. Journal of Ethnopharmacology, 271: 113877.
doi: 10.1016/j.jep.2021.113877 |
| [17] |
GOLLAN P J, TIKKANEN M, ARO E M, 2015. Photosynthetic light reactions: integral to chloroplast retrograde signalling[J]. Current Opinion in Plant Biology, 27: 180-191.
doi: 10.1016/j.pbi.2015.07.006 pmid: 26318477 |
| [18] |
GONG ZHIZHONG, DONG CHUNHAI, LEE H, et al, 2005. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis[J]. The Plant Cell, 17(1): 256-267.
doi: 10.1105/tpc.104.027557 |
| [19] |
JACOBSEN S E, RUNNING M P, MEYEROWITZ E M, 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems[J]. Development, 126(23): 5231-5243.
doi: 10.1242/dev.126.23.5231 pmid: 10556049 |
| [20] |
KIM J S, KIM K A, OH T R, et al, 2008. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions[J]. Plant Cell Physiol, 49(10): 1563-71.
doi: 10.1093/pcp/pcn125 pmid: 18725370 |
| [21] |
KÖHLER D, SCHMIDT-GATTUNG S, BINDER, 2010. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana[J]. Plant Mol Biol, 72(4-5): 459-467.
doi: 10.1007/s11103-009-9584-9 |
| [22] |
KREPS JA, WU Y, CHANG HS, ZHU T, et al, 2002. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress[J]. Plant Physiol, 130(4): 2129-2141.
pmid: 12481097 |
| [23] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [24] |
LI D, LIU H, ZHANG H, et al, 2008. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress[J]. J Exp Bot, 59(8): 2133-2146.
doi: 10.1093/jxb/ern072 pmid: 18441339 |
| [25] |
LI S C, CHUNG M C, CHEN C S, 2001. Cloning and characterization of a DEAD box RNA helicase from the viable seedlings of aged mung bean[J]. Plant Mol Biol, 47(6): 761-770.
doi: 10.1023/a:1013687412020 pmid: 11785937 |
| [26] | LINDER P, JANKOWSKY E, 2011. From unwinding to clamping the DEAD box RNA helicase family[J]. Nat Rev Mol Cell Biol, 12(8): 505-516. |
| [27] |
LINDER P, 2006. Dead-box proteins: a family affairactive and passive players in RNP-remodeling[J]. Nucleic Acids Res, 34(15): 4168-80.
doi: 10.1093/nar/gkl468 |
| [28] |
LIU Y, TABATA D, IMAI R, 2016. A cold-inducible DEAD-Box RNA Helicase from Arabidopsis thaliana regulates plant growth and development under low temperature[J]. PLoS One, 11(4): e0154040.
doi: 10.1371/journal.pone.0154040 |
| [29] |
LIVAK K J, SCHMITTGEN T D, 2001. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2-ΔΔCT method[J]. Methods, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 |
| [30] |
LORKOVIC ZJ, HERMANN RG, OELMULLER R, 1997. PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants[J]. Mol Cell Biol, 17(4): 2257-2265.
doi: 10.1128/MCB.17.4.2257 pmid: 9121476 |
| [31] |
LUO Y, LIU YB, DONG YX, et al, 2009. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment[J]. J Plant Physiol, 166(4): 385-394.
doi: 10.1016/j.jplph.2008.06.018 |
| [32] |
MATTES A, SCHMIDT-GATTUND S, KOHLER D, et al, 2007. Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria[J]. Plant Physiol, 145(4): 1637-1646.
doi: 10.1104/pp.107.108076 pmid: 17951454 |
| [33] |
MAZROUI R, SUKARIEH R, BORDELEAU ME, et al, 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation[J]. Mol Biol Cell, 17(10): 4212-4219.
pmid: 16870703 |
| [34] |
MATTHES A, SCHMIDT-GATTUNG S, KOHLER D, et al, 2007. Two DEAD-box proteins may be part of RNA-dependent high-molecularmass protein complexes in Arabidopsis mitochondria[J]. Plant Physiol, 145(4): 1637-1646.
doi: 10.1104/pp.107.108076 |
| [35] |
NAWAZ G, LEE K, PARK S J, et al, 2018a. A chloroplast-targeted cabbage DEAD-box RNA helicase BrRH22 confers abiotic stress tolerance to transgenic Arabidopsis plants by affecting translation of chloroplast transcripts[J]. Plant Physiology and Biochemistry, 127: 336-342.
doi: 10.1016/j.plaphy.2018.04.007 |
| [36] |
NAWAZ G, SAI T Z T, LEE K, et al, 2018b. Rice DEAD-box RNA helicase OsRH53 has negative impact on Arabidopsis response to abiotic stresses[J]. Plant Growth Regulation, 85(1): 153-163.
doi: 10.1007/s10725-018-0381-9 |
| [37] |
NAWAZ G, KANG H, 2019. Rice OsRH58, a chloroplast DEAD-box RNA helicase, improves salt or drought stress tolerance in Arabidopsis by affecting chloroplast translation[J]. BMC Plant Biol, 19(1): 17.
doi: 10.1186/s12870-018-1623-8 |
| [38] |
NIDUMUKKALA S, TAYI L, CHITTELA R K, et al, 2019. DEAD box helicases as promising molecular tools for engineering abiotic stress tolerance in plants[J]. Critical Reviews in Biotechnology, 39(3): 395-407.
doi: 10.1080/07388551.2019.1566204 pmid: 30714414 |
| [39] |
NISHIMURA K, ASHIDA H, OGAWA T, et al, 2010. A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA[J]. The Plant Journal, 63(5): 766-777.
doi: 10.1111/j.1365-313X.2010.04276.x pmid: 20561259 |
| [40] |
OBERSCHELP G P J, GUARNASCHELLI A B, TESON N, et al, 2020. Cold acclimation and freezing tolerance in three Eucalyptus species: a metabolomic and proteomic approach[J]. Plant Physiology and Biochemistry, 154: 316-327.
doi: 10.1016/j.plaphy.2020.05.026 |
| [41] |
OP DEN CAMP R, KUHLEMEIER C, 1998. Phosphorylation of tobacco eukaryotic translation initiation factor 4A upon pollen tube germination[J]. Nucleic Acids Research, 26(9): 2058-2062.
pmid: 9547259 |
| [42] |
REN QIFEI, ZHOU YUNCHAO, ZHOU XINWEI, 2020. Combined transcriptome and proteome analysis of Masson Pine (Pinus massoniana Lamb.) seedling root in response to nitrate and ammonium supplementations[J]. International Journal of Molecular Sciences, 21(2): 7548.
doi: 10.3390/ijms21207548 |
| [43] |
SEKI M, NARUSAKA M, ABE H, et al, 2001. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray[J]. The Plant Cell, 13(1): 61-72.
doi: 10.1105/tpc.13.1.61 |
| [44] |
STONEBLOOM S, BURCH-SMITH T, KIM I, et al, 2009. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata[J]. Proceedings of the National Academy of Sciences of the United States of America, 106(40): 17229-17234.
doi: 10.1073/pnas.0909229106 pmid: 19805190 |
| [45] |
TANNER N K, LINDER P, 2001. DExD/H box RNA helicases[J]. Molecular Cell, 8(2): 251-262.
doi: 10.1016/S1097-2765(01)00329-X |
| [46] | TUTEJA N, SAHOO R K, GARG B, et al, 2013. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64)[J]. The Plant Journal, 76(1): 115-127. |
| [47] |
VASHISHT A A, PRADHAN A, TUTEJA R, et al, 2005. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C[J]. The Plant Journal, 44(1): 76-87.
doi: 10.1111/j.1365-313X.2005.02511.x |
| [48] |
VASHISHT A A, TUTEJA N, 2006. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance[J]. Journal of Photochemistry and Photobiology B: Biology, 84(2): 150-160.
doi: 10.1016/j.jphotobiol.2006.02.010 |
| [49] |
WEBSTER C, GAUT R L, BROWNING K S, et al, 1991. Hypoxia enhances phosphorylation of eukaryotic initiation factor 4A in maize root tips[J]. Journal of Biological Chemistry, 266(34): 23341-23346.
pmid: 1744128 |
| [50] |
WESTERN T L, CHENG YULAN, LIU JUN, et al, 2002. HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis[J]. Development, 129(7): 1569-1581.
doi: 10.1242/dev.129.7.1569 |
| [51] |
ZHANG BIN, JIA DONG, GAO ZHIQIANG, et al, 2016. Physiological responses to low temperature in spring and winter wheat varieties[J]. Journal of the Science of Food and Agriculture, 96(6): 1967-1973.
doi: 10.1002/jsfa.7306 pmid: 26095741 |
| [52] |
ZHU MINGKU, CHEN GUOPING, DONG TINGTING, et al, 2015. SlDEAD31, a putative DEAD-box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato[J]. PLoS One, 10(8): e0133849.
doi: 10.1371/journal.pone.0133849 |
| [1] | 吴鸿博, 罗锋, 陈治澎, 朱飞, 曾靖伟, 张弛, 李瑞杰. 红树林生态重建效果预测研究新模式[J]. 热带海洋学报, 2024, 43(4): 86-97. |
| [2] | 郑法, 黄福林, 陈泽恒, 丁伟品. 基于LUCC和景观格局变化的广西山口红树林湿地动态研究[J]. 热带海洋学报, 2024, 43(4): 165-173. |
| [3] | 周治刚, 岳文, 李辉权, 林阳阳. 树种类型和潮滩高程对广东湛江高桥红树林碳储量的影响[J]. 热带海洋学报, 2024, 43(2): 108-120. |
| [4] | 申键, 简焯锴, 欧阳雪敏, 艾彬. 结合潮位校正的雷州半岛红树林湿地动态变迁遥感监测[J]. 热带海洋学报, 2024, 43(1): 137-153. |
| [5] | 耿婉璐, 邢永泽, 张秋丰, 管卫兵. 广西北海红树林宜林滩涂大型底栖动物群落结构特征[J]. 热带海洋学报, 2024, 43(1): 107-115. |
| [6] | 董俊德, 黄小芳, 龙爱民, 王友绍, 凌娟, 杨清松. 红树林固氮微生物及其生态功能研究进展[J]. 热带海洋学报, 2023, 42(4): 1-11. |
| [7] | 梁寒峭, 陈文凤, 范益铠, 朱子冬, 马国需, 陈德力, 田婧. 红树林来源曲霉属和木霉属内生真菌次生代谢产物及活性研究进展[J]. 热带海洋学报, 2023, 42(4): 12-24. |
| [8] | 张程飞, 任广波, 吴培强, 胡亚斌, 马毅, 阎宇, 张菁锐. 基于高分光学与全极化SAR的海南八门湾红树林种间分类方法[J]. 热带海洋学报, 2023, 42(2): 153-168. |
| [9] | 周月月, 王友绍. 广东沿海红树林区水质变化特征与富营养状态评估[J]. 热带海洋学报, 2022, 41(6): 1-11. |
| [10] | 吴伟志, 赵志霞, 杨升, 梁立成, 陈秋夏, 卢翔, 刘星, 张小伟. 浙江省红树林分布和造林成效分析[J]. 热带海洋学报, 2022, 41(6): 67-74. |
| [11] | 谢勇, 王友绍, 张维仕. 红海榄肉桂酸-4-羟基化酶基因的克隆与表达分析[J]. 热带海洋学报, 2022, 41(6): 20-27. |
| [12] | 李华薇, 徐向荣. 中国典型红树林沉积物中多溴联苯醚和替代型溴系阻燃剂污染特征[J]. 热带海洋学报, 2022, 41(1): 117-130. |
| [13] | 戴志军, 周晓妍, 王杰, 胡宝清. 红树林潮滩沉积动力研究进展与展望[J]. 热带海洋学报, 2021, 40(3): 69-75. |
| [14] | 王友绍. 全球气候变化对红树林生态系统的影响、挑战与机遇[J]. 热带海洋学报, 2021, 40(3): 1-14. |
| [15] | 董迪, 曾纪胜, 魏征, 严金辉. 联合星载光学和SAR影像的漳江口红树林与互花米草遥感监测[J]. 热带海洋学报, 2020, 39(2): 107-117. |
|
||