热带海洋学报 ›› 2023, Vol. 42 ›› Issue (2): 54-63.doi: 10.11978/2022083CSTR: 32234.14.2022083
海南澳洲管体星虫线粒体基因组特征及进化分析
黄培贤1,2( ), 姚雪梅1,2(
), 姚雪梅1,2( ), 余巧驰1,2, 张佳玉1,2
), 余巧驰1,2, 张佳玉1,2
- 1.南海海洋资源利用国家重点实验室(海南大学), 海南 海口 570228
2.海南大学海洋学院, 海南 海口 570228
-
收稿日期:2022-04-19修回日期:2022-07-22出版日期:2023-03-10发布日期:2022-07-27 -
通讯作者:姚雪梅。email:yaoxuemei72@163.com -
作者简介:黄培贤(1997—), 海南省琼海市人, 在读硕士研究生, 从事海洋生物学方面研究。email: 85200494@qq.com
-
基金资助:海南省自然科学基金(319MS013)
Mitogenome characteristics and phylogenetic analysis of Siphonosoma australe in Hainan
HUANG Peixian1,2( ), YAO Xuemei1,2(
), YAO Xuemei1,2( ), YU Qiaochi1,2, ZHANG Jiayu1,2
), YU Qiaochi1,2, ZHANG Jiayu1,2
- 1. State Key Laboratory of Marine Resource Utilization in South China Sea, Hainan University, Haikou 570228, China
2. College of Ocean, Hainan University, Haikou 570228, China
-
Received:2022-04-19Revised:2022-07-22Online:2023-03-10Published:2022-07-27 -
Contact:YAO Xuemei. email:yaoxuemei72@163.com -
Supported by:Hainan Provincial Natural Science Foundation of China(319MS013)
摘要:
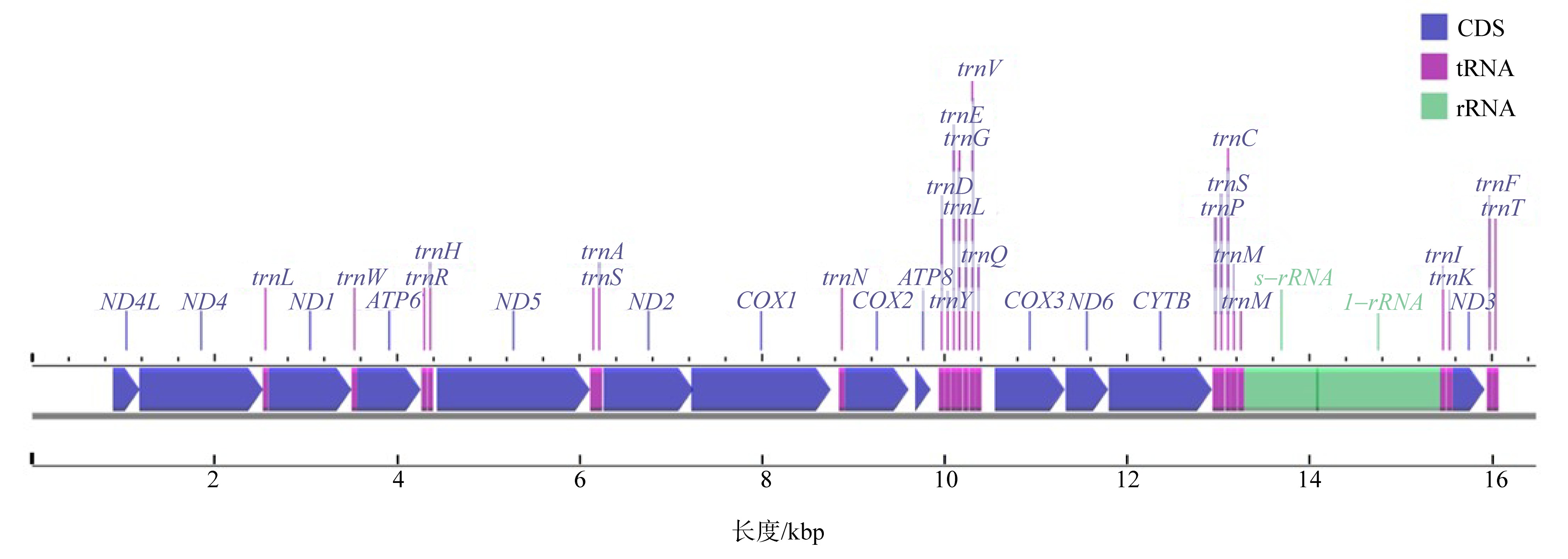
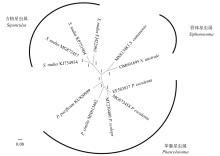
澳洲管体星虫是海南当地的特色海产资源, 隶属管体星虫属。该属在星虫动物门内的分类地位一直极具争议。文章通过高通量测序测定海南文昌地区澳洲管体星虫的线粒体基因组, 与GenBank中收集的星虫线粒体基因组进行比对分析, 解析其基因组序列特征, 并进一步探讨管体星虫属在星虫动物门内的进化地位。结果显示: 澳洲管体星虫线粒体基因组长度为16483bp, 包含38个基因(13个蛋白质编码基因、23个tRNAs和2个rRNAs)。线粒体基因组呈AT偏好, 其A+T的含量为65.87%。分析相对同义密码子使用度发现, 澳洲管体星虫线粒体蛋白质编码基因对结尾为A和U的密码子具有明显偏好性。澳洲管体星虫线粒体蛋白编码基因COX1、COX3、ND5的氨基酸数量与其他星虫比较, 差异较大。澳洲管体星虫与GenBank中收集的星虫线粒体基因组的主编码基因中, COX1、COX2和CYTB基因变异位点比例低, 而ND2、ND4L和ND6基因变异位点比例高, ATP8基因的变异位点的比例最高(83.33%)。采用邻接法(neighbor-joining, NJ)、最大似然法(maximum likelihood, ML)、贝叶斯法(Bayesian inference, BI), 利用线粒体基因组核酸序列构建进化树, 结果显示, 管体星虫属介于革囊星虫属和方格星虫属之间, 与革囊星虫属关系较近, 反而与方格星虫属的关系较远, 与传统的形态学分类不一致。形态学上, 管体星虫属混合了革囊星虫属和方格星虫属的形态特征, 而线粒体基因组构建的进化树更能体现管体星虫属真实且独特的进化地位。
引用本文
黄培贤, 姚雪梅, 余巧驰, 张佳玉. 海南澳洲管体星虫线粒体基因组特征及进化分析[J]. 热带海洋学报, 2023, 42(2): 54-63.
HUANG Peixian, YAO Xuemei, YU Qiaochi, ZHANG Jiayu. Mitogenome characteristics and phylogenetic analysis of Siphonosoma australe in Hainan[J]. Journal of Tropical Oceanography, 2023, 42(2): 54-63.
表1
外源信息表"
| 属 | 物种 | 采样地 | GenBank 登录号 |
|---|---|---|---|
| 方格星虫属Sipunculus | 光裸方格星虫(Sipunculus nudus) | Concarneau, France | FJ422961 |
| Beibu Bay, China | MG873457 | ||
| Gulei, China | KJ754934 | ||
| Yantai, China | KP751904 | ||
| 革囊星虫属Phascolosoma | 可口革囊星虫(Phascolosoma esculenta) | Beibu Bay, China | MG873458 |
| Wenzhou, China | EF583817 | ||
| 太平洋革囊星虫(Phascolosoma pacificum) | Chuuk, Micronesia | KU820989 | |
| 厥目革囊星虫(Phascolosoma scolops) | Beibu Bay, China | MT239480 | |
| 类革囊星虫(Phascolosoma similis) | Beibu Bay, China | MN813482 | |
| 管体星虫属Siphonosoma | 库岛管体星虫(Siphonosoma cumanense) | Beibu Bay, China | MN813483 |
表3
澳洲管体星虫线粒体蛋白质编码基因的相对同义密码子使用度(RSCU)"
| 氨基酸 | 密码子 | 次数 | RSCU | 氨基酸 | 密码子 | 次数 | RSCU |
|---|---|---|---|---|---|---|---|
| Phe | UUU | 236 | 1.45 | Tyr | UAU | 91 | 1.35 |
| UUC | 90 | 0.55 | UAC | 44 | 0.65 | ||
| Leu | UUA | 226 | 2.17 | His | CAU | 56 | 1.30 |
| UUG | 20 | 0.19 | CAC | 30 | 0.70 | ||
| CUU | 173 | 1.66 | Gln | CAA | 62 | 1.70 | |
| CUC | 68 | 0.65 | CAG | 11 | 0.30 | ||
| CUA | 127 | 1.22 | Asn | AAU | 71 | 1.31 | |
| CUG | 10 | 0.10 | AAC | 37 | 0.69 | ||
| Ile | AUU | 226 | 1.52 | Lys | AAA | 79 | 1.82 |
| AUC | 71 | 0.48 | AAG | 8 | 0.18 | ||
| Met | AUA | 161 | 1.71 | Asp | GAU | 34 | 1.05 |
| AUG | 27 | 0.29 | GAC | 31 | 0.95 | ||
| Val | GUU | 78 | 1.70 | Glu | GAA | 63 | 1.70 |
| GUC | 25 | 0.55 | GAG | 11 | 0.30 | ||
| GUA | 73 | 1.60 | Cys | UGU | 24 | 1.33 | |
| GUG | 7 | 0.15 | UGC | 12 | 0.67 | ||
| Ser | UCU | 117 | 2.77 | Trp | UGA | 87 | 1.81 |
| UCC | 41 | 0.97 | UGG | 9 | 0.19 | ||
| UCA | 84 | 1.99 | Arg | CGU | 22 | 1.22 | |
| UCG | 7 | 0.17 | CGC | 11 | 0.61 | ||
| AGU | 19 | 0.45 | CGA | 33 | 1.83 | ||
| AGC | 13 | 0.31 | CGG | 6 | 0.33 | ||
| AGA | 51 | 1.21 | Pro | CCU | 71 | 1.41 | |
| AGG | 6 | 0.14 | CCC | 29 | 0.57 | ||
| Thr | ACU | 93 | 1.52 | CCA | 95 | 1.88 | |
| ACC | 38 | 0.62 | CCG | 7 | 0.14 | ||
| ACA | 102 | 1.67 | Gly | GGU | 44 | 0.91 | |
| ACG | 12 | 0.20 | GGC | 28 | 0.58 | ||
| Ala | GCU | 96 | 1.39 | GGA | 98 | 2.02 | |
| GCC | 53 | 0.77 | GGG | 24 | 0.49 | ||
| GCA | 115 | 1.67 | |||||
| GCG | 12 | 0.17 |
表4
星虫动物3个属7个种的线粒体蛋白质编码基因的氨基酸数量"
| 星虫名称 | ATP6 | ATP8 | CYTB | COX1 | COX2 | COX3 | ND1 | ND2 | ND3 | ND4 | ND4L | ND5 | ND6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 可口革囊星虫 | 231 | 54 | 378 | 519 | 231 | 267 | 304 | 323 318 | 121 | 450 451 | 94 | 571 562 | 157 |
| 太平洋革囊星虫 | 231 | 49 | 378 | 518 | 227 | 263 | 308 | 319 | 118 | 450 | 94 | 572 | 157 |
| 类革囊星虫 | 231 | 53 | 378 | 519 | 230 | 267 | 303 | 327 | 121 | 450 | 94 | 571 | 157 |
| 厥目革囊星虫 | 231 | 54 | 378 | 519 | 229 | 291 | 304 | 320 | 121 | 451 | 94 | 572 | 157 |
| 光裸方格星虫 | 229 234 | 52 53 | 379 | 516 519 | 231 230 | 259 | 314 312 | 328 327 329 | 119 117 | 452 454 | 93 | 565 571 | 157 |
| 库岛管体星虫 | 231 | 54 | 379 | 513 | 231 | 275 | 303 | 318 | 114 | 448 | 94 | 569 | 155 |
| 澳洲管体星虫 | 231 | 56 | 377 | 510 | 232 | 293 | 303 | 327 | 116 | 452 | 94 | 559 | 155 |
表5
星虫动物3个属7个种的线粒体基因组的主编码基因变异位点分析"
| 基因名称 | 总位点数 | 保守位点数 | 变异位点数 | 变异位点比例/% |
|---|---|---|---|---|
| ATP6 | 669 | 226 | 443 | 66.22 |
| ATP8 | 144 | 24 | 120 | 83.33 |
| CYTB | 1131 | 550 | 581 | 51.37 |
| COX1 | 1531 | 824 | 707 | 46.18 |
| COX2 | 680 | 328 | 352 | 51.76 |
| COX3 | 780 | 342 | 438 | 56.15 |
| ND1 | 906 | 368 | 538 | 59.38 |
| ND2 | 944 | 215 | 729 | 77.22 |
| ND3 | 344 | 126 | 218 | 63.37 |
| ND4 | 1329 | 402 | 927 | 69.75 |
| ND4L | 282 | 77 | 205 | 72.70 |
| ND5 | 1657 | 523 | 1134 | 68.44 |
| ND6 | 466 | 118 | 348 | 74.68 |
| 12S rRNA | 753 | 295 | 458 | 60.82 |
| 16S rRNA | 980 | 423 | 557 | 56.84 |
| [1] |
陈子安, 杜晓东, 王庆恒, 等, 2007. 3种星虫线粒体16S rRNA、COI和Cytb基因片段的序列比较[J]. 广东海洋大学学报, 27(4): 3-10.
|
|
|
|
| [2] |
傅静, 孙啸, 2003. 基于全基因组的系统发生分析[J]. 生物技术, (6): 53-56.
|
|
|
|
| [3] |
黄族豪, 刘迺发, 2010. 动物线粒体基因组变异研究进展[J]. 生命科学研究, 14(2): 166-171.
|
|
|
|
| [4] |
蒋文枰, 李家乐, 郑润玲, 等, 2010. 褶纹冠蚌线粒体基因组全序列分析[J]. 遗传, 32(2): 153-162.
|
|
|
|
| [5] |
兰国宝, 杨素芳, 谢体三, 等, 2007. 星虫动物门系统发生研究进展[J]. 广西科学, 14(2): 186-192.
|
|
|
|
| [6] |
李凤鲁, 周红, 王玮, 1992. 中国沿海管体星虫属(星虫动物门)的研究[J]. 青岛海洋大学学报: 自然科学版, 22(1): 97-102.
|
|
|
|
| [7] |
孟乾, 张志勇, 张志伟, 等, 2020. 斑石鲷和条石鲷线粒体基因组密码子使用分析[J]. 水产科学, 39(5): 702-709.
|
|
|
|
| [8] |
彭银辉, 周于娜, 刘旭佳, 等, 2017. 基于线粒体控制区序列的光裸方格星虫群体遗传多样性分析[J]. 水产学报, 41(10): 1542-1551.
|
|
|
|
| [9] |
乔立君, 姚雪梅, 余巧驰, 2022. 海南澳洲管体星虫卵细胞发育及生殖周期研究[J]. 热带海洋学报, 41(5): 161-169.
doi: 10.11978/2021143 |
|
|
|
| [10] |
申欣, 2012. 星虫动物线粒体基因组全序列的基因排列和特征比较[J]. 水产科学, 31(9): 554-559.
|
|
|
|
| [11] |
宋素霞, 2015. 中国沿海光裸方格星虫线粒体基因组分析及遗传多样性研究[D]. 福建: 集美大学.
|
|
|
|
| [12] |
宋素霞, 丁少雄, 鄢庆枇, 等, 2017. 中国沿海光裸方格星虫6个地理群体遗传多样性分析[J]. 水产学报, 41(8): 1183-1191.
|
|
|
|
| [13] |
孙萌, 2021. 基于多基因对寡膜纲纤毛虫重要类群分子系统学的研究[D]. 黑龙江: 哈尔滨师范大学.
|
|
|
|
| [14] |
夏立萍, 徐鸣, 郭宝英, 等, 2021. 等边浅蛤线粒体全基因组测序和系统发育研究[J]. 浙江海洋大学学报(自然科学版), 40(2): 93-100.
|
|
|
|
| [15] |
于黎, 张亚平, 2006. 系统发育基因组学——重建生命之树的一条迷人途径[J]. 遗传, 28(11): 1445-1450.
|
|
|
|
| [16] |
钟声平, 蒋艳, 刘永宏, 等, 2020. 广西5种常见星虫动物的线粒体基因组特征和系统进化分析[J]. 广西科学, 27(5): 532-540.
|
|
|
|
| [17] |
周红, 李凤鲁, 王玮, 2007. 中国动物志, 无脊椎动物. 第四十六卷, 星虫动物门螠虫动物门[M]. 北京: 科学出版社.
|
|
|
|
| [18] |
doi: 10.1093/genetics/159.2.623 pmid: 11606539 |
| [19] |
|
| [20] |
doi: 10.2307/2413324 |
| [21] |
doi: 10.1007/s00227-010-1402-z |
| [22] |
doi: 10.1111/maec.12104 |
| [23] |
doi: 10.1111/j.1463-6409.2011.00507.x |
| [24] |
doi: 10.1016/j.ympev.2014.10.019 pmid: 25450098 |
| [25] |
doi: 10.1093/bioinformatics/btp187 pmid: 19346325 |
| [26] |
|
| [27] |
pmid: 12470944 |
| [28] |
doi: 10.1186/1471-2164-10-27 |
| [29] |
doi: 10.1093/sysbio/sys029 pmid: 22357727 |
| [30] |
|
| [31] |
doi: 10.1186/1471-2164-10-136 |
| [32] |
doi: 10.1016/j.gene.2007.06.021 |
| [33] |
doi: 10.1111/ivb.2003.122.issue-3 |
| [34] |
doi: 10.1093/molbev/msr121 pmid: 21546353 |
| [35] |
doi: 10.1007/978-1-59745-581-7_12 pmid: 18629668 |
| [36] |
doi: 10.1080/23802359.2020.1731378 |
| [1] | 赵中伟, 吴凌云, 高伟健, 李伟. 海南岛峨蔓五彩湾内海蚀平台对海蚀崖侵蚀的影响研究[J]. 热带海洋学报, 2024, 43(5): 106-115. |
| [2] | 李子若, 罗晏杰, 曹政, CHIN Yaoxian, 王沛政. 短指软珊瑚(Sinularia acuta)热休克蛋白HSP70家族特征及进化分析[J]. 热带海洋学报, 2024, 43(4): 123-136. |
| [3] | 霍嘉欣, 李颖心, 宋严, 朱晴, 周伟华, 袁翔城, 黄晖, 刘胜. 木珊瑚科Cladopsammia gracilis和Rhizopsammia wettsteini 线粒体全基因组比较与系统进化分析*[J]. 热带海洋学报, 2024, 43(3): 22-30. |
| [4] | 曾维特, 张东强, 刘兵, 杨永鹏, 张航飞, 吴多誉, 王晓林. 海南岛北部海湾表层海水重金属分布特征、主控因素及污染评价[J]. 热带海洋学报, 2023, 42(6): 156-167. |
| [5] | 乔立君, 姚雪梅, 余巧驰. 海南澳洲管体星虫卵细胞发育及生殖周期研究[J]. 热带海洋学报, 2022, 41(5): 161-169. |
| [6] | 毛颖津, 高贝乐. 比较基因组学分析弧菌属群体感应通路的分布与进化[J]. 热带海洋学报, 2022, 41(1): 28-41. |
| [7] | 刘金梅, 黄冰心, 丁兰平, 王雪聪, 闫璟, 闫盼竹, 张瑶. 我国海南省东南部台湾那屿藻的形态观察及系统发育分析[J]. 热带海洋学报, 2021, 40(6): 76-82. |
| [8] | 王媛琪, 杨阳, 周亮, 汪亚平, 高抒. 基于风暴水流能量分布对海南岛小东海珊瑚巨砾成因的分析*[J]. 热带海洋学报, 2021, 40(4): 110-121. |
| [9] | 洪义国, 焦黎静, 吴佳鹏, 龙爱民, 王伟. 海洋亚硝酸盐氧化细菌的多样性分布及其生态功能研究进展[J]. 热带海洋学报, 2021, 40(2): 139-146. |
| [10] | 孟天, 陈佐, 朱军, 邹潇潇, 符青艳, 鲍时翔. 海南岛海洋绿藻新记录种未氏仙掌藻(Halimeda velasquezii)的形态学与分子系统学分析[J]. 热带海洋学报, 2020, 39(6): 114-121. |
| [11] | 杨敏, 孔晓瑜, 时伟, 龚理. 长臂缨鲆核糖体RNA基因序列多态性特征分析[J]. 热带海洋学报, 2019, 38(1): 55-66. |
| [12] | 王天宇, 杜岩, 夏一凡. 基于拉格朗日方法的南海自动剖面浮标轨迹模拟系统*[J]. 热带海洋学报, 2018, 37(4): 9-9. |
| [13] | 李永梅, 刘瑞, 杨楠, 丁兰平, 黄冰心, 王艺晓, 陈彦伟. 海南省4种江蓠属(红藻门)海藻的形态分类学研究[J]. 热带海洋学报, 2018, 37(4): 29-37. |
| [14] | 冯兴如, 李近元, 尹宝树, 杨德周, 陈海英, 高冠东. 海南东方近岸海域海浪观测特征研究[J]. 热带海洋学报, 2018, 37(3): 1-8. |
| [15] | 廖秀丽, 戴明, 巩秀玉, 刘华雪, 黄洪辉. 南海南部次表层叶绿素a质量浓度最大值及其影响因子*[J]. 热带海洋学报, 2018, 37(1): 45-56. |
|
||