Journal of Tropical Oceanography ›› 2025, Vol. 44 ›› Issue (6): 120-131.doi: 10.11978/2025022CSTR: 32234.14.2025022
Previous Articles Next Articles
Identification and structural analysis of the HicAB toxin-antitoxin system encoded by a prophage in coral-associated Halomonas meridiana
ZHANG Yu1,2( ), LIU Ziyao2, WANG Xiaoxue2, CHEN Ran2(
), LIU Ziyao2, WANG Xiaoxue2, CHEN Ran2( )
)
- 1
College of Life Science and Technology ,Jinan University
2Key Laboratory of Tropical Marine Bio-resources and Ecology ,South China Sea Institute of Oceanology, Chinese Academy of Sciences
-
Received:2025-02-17Revised:2025-03-14Online:2025-11-10Published:2025-12-03 -
Contact:CHEN Ran. email: chenran@scsio.ac.cn -
Supported by:National Natural Science Foundation of China(42188102); Special Fund of the South China Sea Institute of Oceanology, Chinese Academy of Sciences(SCSIO2023QY03); Ocean Negative Carbon Emissions Program(ONCE); Open project of State Key Laboratory of South China Sea Marine Resources Utilization, Hainan University(MRUKF2023001)
CLC Number:
- Q939.9
Cite this article
ZHANG Yu, LIU Ziyao, WANG Xiaoxue, CHEN Ran. Identification and structural analysis of the HicAB toxin-antitoxin system encoded by a prophage in coral-associated Halomonas meridiana[J].Journal of Tropical Oceanography, 2025, 44(6): 120-131.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Bacterial strains used in this study"
| 菌株 | 描述 | 来源 |
|---|---|---|
| Hm43005 | Isolated from Coral samples of G. fascicularis. | 实验室保存 |
| WM3064 | thrB1004 pro thi rpsL hsdS lacZΔM15 RP4-360 Δ(araBAD)567 ΔdapA1341:: [erm pir] | 实验室保存 |
| BL21 (DE3) | F-ompT hsdSB(rB-mB-) gal dcm λ(DE3) Ω PtacUV5::T7 polymerase | Novagen |
| BW25113 (K-12) | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 rph-1 | Baba et al, 2006 |
| Top10 | F-, mcrAΔ(mrr-hsd RMS-mcrBC), ϕ80, lacZΔM15, ΔlacX74, recA1, araΔ139Δ(ara-leu)7697, galU, galK, rps, (Strr) endA1, nupG | 实验室保存 |
Tab. 2
Plasmids used in this study"
| 质粒 | 描述 | 来源 |
|---|---|---|
| pHGECm | CmR, IPTG inducible expression vector in E.coli and H.meridiana SCSIO 43005 strains | 实验室保存 |
| pHGECm-hicA | CmR; over-expression vector for CTT34_05265 | 本研究 |
| pHGECm-hicB | CmR; over-expression vector for CTT34_05260 | 本研究 |
| pHGECm-hicAB | CmR; over-expression vector for CTT34_05260 and CTT34_05265 as one operon | 本研究 |
| pUT18C-hicA | AmpR; interaction assay vector for CTT34_05265 | 本研究 |
| pKT25-hicB | KmR; interaction assay vector for CTT34_05260 | 本研究 |
| pET28b | KmR, IPTG inducible expression and purified vector | Novagen |
| pET28b-hicA-his | KmR; pET28b PT7-lac::hicA with C-terminal His-tagged | 本研究 |
| pET28b-hicB-his | KmR; pET28b PT7-lac::hicB with C-terminal His-tagged | 本研究 |
| pET28b-his-hicAB | KmR; pET28b PT7-lac:: hicAB with N-terminal His-tagged | 本研究 |
Tab. 3
Sequences of designed primers used in this study"
| 引物名称 | 序列(5′-3′) | 用途 |
|---|---|---|
| pHGECm-hicA-F | TAACAATTTCACACAGGAGAGATGAACAGCAGAGCACTGATC | 毒性验证 |
| pHGECm-hicA-R | ATCCGCCAAAACAGCCAAGCTTCATTCGAGGCCAGCGCTTTTC | 毒性验证 |
| pHGECm-hicB-F | TAACAATTTCACACAGGAGAGATGTTATTTCCCATTGCCATTG | 毒性验证 |
| pHGECm-hicB-R | ATCCGCCAAAACAGCCAAGCTTTAGCTGGCTTGTTTGTTCCTG | 毒性验证 |
| pUT18C-hicA-F | ACTCTAGAGGATCCCCGGGTACCGATGAACAGCAGAGCACTGATC | 细菌双杂交 |
| pUT18C-hicA-R | ATTACTTAGTTATATCGATGAATTTCATTCGAGGCCAGCGCTTTTC | 细菌双杂交 |
| pKT25-hicB-F | CTAGAGGATCCCCGGGTACCTATGTTATTTCCCATTGCCATTG | 细菌双杂交 |
| pKT25-hicB-R | GAATTCTTAGTTACTTAGTTAGCTGGCTTGTTTGTTCCTG | 细菌双杂交 |
| pET28b-hicA-his-F | CTTTAAGAAGGAGATATACCATGAACAGCAGAGCACTGATC | 蛋白表达纯化 |
| pET28b-hicA-his-R | CTCGAGTGCGGCCGCAAGCTTTTCGAGGCCAGCGCTTTTCC | 蛋白表达纯化 |
| pET28b-hicB-his-F | CTTTAAGAAGGAGATATACCATGTTATTTCCCATTGCCATTG | 蛋白表达纯化 |
| pET28b-hicB-his-R | CTCGAGTGCGGCCGCAAGCTTGCTGGCTTGTTTGTTCCTGGC | 蛋白表达纯化 |
| pET28b-his-hicAB-F | GGTGGACAGCAAATGGGTCGGATGAACAGCAGAGCACTGATC | Pull-down分析 |
| pET28b-his-hicAB-R | CTCGAGTGCGGCCGCAAGCTTTTAGCTGGCTTGTTTGTTCCTG | Pull-down分析 |

Fig. 1
Bioinformatics analysis of the HicAB system. (a) The location of hicAB operon within the prophage, with hicA and hicB shown in blue and green, respectively; (b) sequence alignment of HmHicA encoded by the Hm43005 strain with the structurally resolved HicA sequences from B. pseudomallei, Y. pestis, S. pneumoniae, and E. coli; (c) sequence alignment of HmHicB encoded by the Hm43005 strain with the structurally resolved HicB sequences from B. pseudomallei, Y. pestis, E. coli and S. pneumoniae"

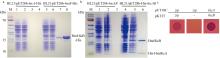
Fig. 3
Experimental validation of the interaction between HmHicA and HmHicB. (a) SDS-PAGE analysis of the size and purity of the purified anti-toxin HmHicB protein: Lane 1 and 5 (pre-induction), Lane 2 and 6 (post-induction), Lane 3 and 7 (elution sample 1), Lane 4 and 8 (elution sample 2); (b) SDS-PAGE analysis demonstrates that the expressed toxin HicA protein successfully pulls down the HmHicB anti-toxin protein: Lane 1 and 4 (pre-induction), Lane 2 and 5 (post-induction), Lane 3 and 6 (elution sample 1); (c) bacterial two-hybrid assay confirms the interaction between HmHicA and HmHicB"
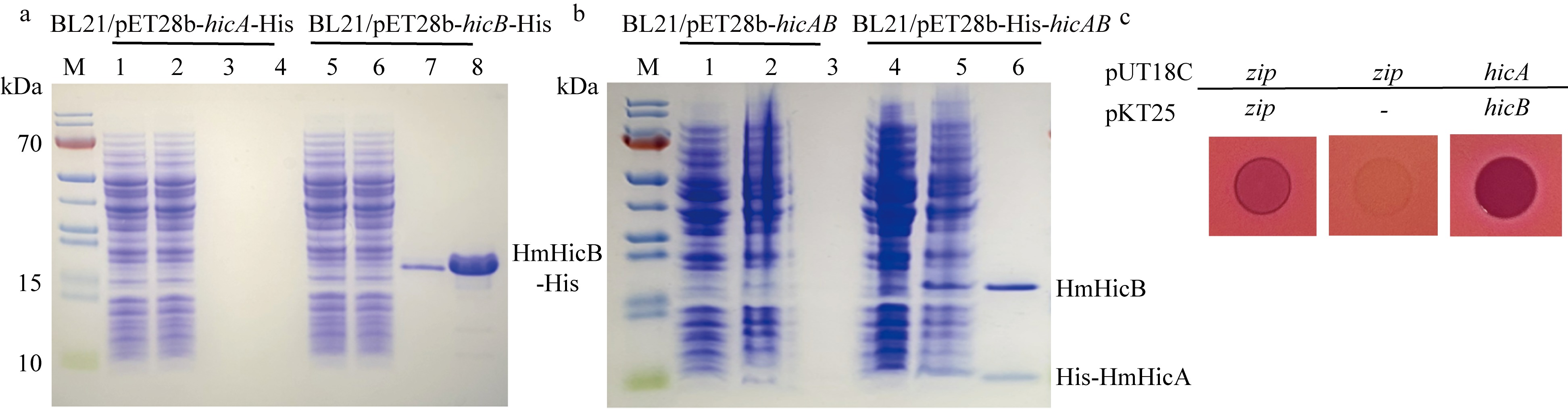

Fig. 4
The in vitro purified HmHicB antitoxin protein can bind to the hicAB promoter. (a) The antitoxin HmHicB can bind to its own promoter, and its binding ability increases with higher protein concentrations. The numbers 0, 1, 2, 4, and 8 represent the molar ratios of protein to nucleic acid (HmHicB/DNA) at 0∶1, 1∶1, 2∶1, 4∶1, and 8∶1, respectively. The control is the promoter of the brnTA system from strain Hm43005. (b) HmHicB binds to the palindromic sequences upstream of the -35 and within the -10 regions of the hicAB promoter. The control is the sequencing result without HmHicB. The -35 and -10 regions are highlighted in red, and the binding regions 1 and 2 are marked with two red dashed boxes. Repeated and palindromic sequences are underlined"

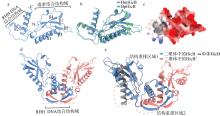
Fig. 6
C-terminal interactions of HmHicB form a typical RHH DNA-binding domain. (a) Monomer structure of HmHicB predicted by AlphaFold 3; (b) comparative structural analysis of HmHicB and B. pseudomallei HicB; (c) surface charge analysis of HmHicB; (d) dimer structure of HmHicB predicted by AlphaFold 3, with the RHH domain formed by dimerization marked by a red dashed box; (e) comparative structural analysis of HmHicB monomer and dimer, with regions undergoing structural rearrangement marked by red dashed ellipses"

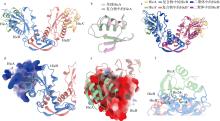
Fig. 7
Structural characterization of the HmHicAB complex. (a) Predicted 2:2 complex structure of HmHicA:HmHicB by AlphaFold 3; (b) structural comparison between the HmHicA monomer and its conformation in the TA complex; (c) structural comparison between the HmHicB dimer and its conformation in the TA complex; (d) surface charge analysis of HmHicA in the TA complex; (e) surface charge analysis of HmHicB in the TA complex; (f) analysis of the surrounding environment of the key active site His24 in HmHicA within the TA complex"

| [1] |
doi: 10.1016/j.molcel.2013.10.014 pmid: 24239291 |
| [2] |
doi: 10.1016/j.ymeth.2012.07.018 |
| [3] |
doi: 10.1093/nar/gkad1220 |
| [4] |
doi: 10.1128/JB.01932-14 |
| [5] |
doi: 10.1093/nar/gkac356 |
| [6] |
doi: 10.1042/BJ20140073 |
| [7] |
doi: 10.1021/acsinfecdis.0c00048 |
| [8] |
doi: S0969-2126(19)30346-6 pmid: 31693911 |
| [9] |
|
| [10] |
doi: 10.1038/nchembio.2078 |
| [11] |
doi: 10.1128/JB.00336-10 |
| [12] |
doi: 10.1093/nar/gkad962 |
| [13] |
doi: 10.1111/emi.2019.21.issue-8 |
| [14] |
doi: 10.1038/s41467-024-51617-x pmid: 39174532 |
| [15] |
doi: 10.3109/10409238.2011.600437 pmid: 21819231 |
| [16] |
doi: 10.1093/nar/gkac387 pmid: 35610055 |
| [17] |
doi: 10.1128/JB.01013-08 |
| [18] |
doi: 10.1038/s41579-021-00661-1 pmid: 34975154 |
| [19] |
doi: 10.1093/nar/gky469 |
| [20] |
doi: 10.1016/j.engmic.2023.100069 |
| [21] |
doi: 10.1093/ismejo/wrae085 |
| [22] |
pmid: 16895922 |
| [23] |
doi: 10.1016/j.str.2019.08.008 |
| [24] |
doi: 10.1111/fml.2012.328.issue-2 |
| [25] |
doi: 10.1002/pro.4792 pmid: 37774136 |
| [26] |
doi: 10.1128/JB.180.17.4693-4703.1998 |
| [27] |
doi: 10.1016/j.cell.2020.09.065 pmid: 33157039 |
| [28] |
|
| [29] |
doi: 10.1038/s41467-018-03652-8 |
| [30] |
doi: 10.1038/srep03186 pmid: 24212724 |
| [31] |
doi: 10.1093/nar/gkab824 pmid: 34551431 |
| [32] |
doi: 10.1093/nar/gkad1011 |
| [33] |
doi: 10.1038/nchembio.560 pmid: 21516113 |
| [34] |
doi: 10.1074/jbc.RA118.005173 pmid: 30337369 |
| [35] |
doi: 10.1093/nar/gkaa706 |
| [36] |
doi: 10.1093/nar/gkaa855 pmid: 33045733 |
| [37] |
pmid: 16741237 |
| [1] | LIN Shituan, WANG Xiaoxue, CHEN Ran. Analysis and identification of seven toxin-antitoxin systems in two cyanobacteria [J]. Journal of Tropical Oceanography, 2022, 41(3): 119-134. |
| [2] | Xuanyu ZHAO, Fei SHI, Xiaoxue WANG. Characterization of a ParE-ParD toxin-antitoxin TA system (SO_A0087-A0088) on a megaplasmid of Shewanella oneidensis [J]. Journal of Tropical Oceanography, 2018, 37(6): 104-111. |
|
||