Journal of Tropical Oceanography ›› 2022, Vol. 41 ›› Issue (1): 52-61.doi: 10.11978/2021018CSTR: 32234.14.2021018
• Marine Biology • Previous Articles Next Articles
Draft genome sequence and comparative genome analysis of Alliroseovarius sp. Z3
LIU Wei1,2( ), GUO Haipeng1,2(
), GUO Haipeng1,2( ), DONG Pengsheng1,2, YAN Mengchen1,2, ZHANG Demin1,2
), DONG Pengsheng1,2, YAN Mengchen1,2, ZHANG Demin1,2
- 1. State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ningbo University, Ningbo 315800, China
2. School of Marine Sciences, Ningbo University, Ningbo 315800, China
-
Received:2021-02-13Revised:2021-04-08Online:2022-01-10Published:2021-04-12 -
Contact:GUO Haipeng E-mail:645560829@qq.com;guohaipeng@nbu.edu.cn -
Supported by:National Natural Science Foundation of China(31672658);General Research Project of Zhejiang Education Department, China(Y201839299);Agricultural Major Project of Ningbo, China(2017C110001)
CLC Number:
- P735.51
Cite this article
LIU Wei, GUO Haipeng, DONG Pengsheng, YAN Mengchen, ZHANG Demin. Draft genome sequence and comparative genome analysis of Alliroseovarius sp. Z3[J].Journal of Tropical Oceanography, 2022, 41(1): 52-61.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
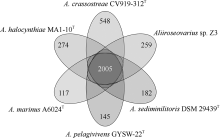
Tab. 1
Average nucleotide identity (ANI) values of six Aliiroseovarius spp."
| Aliiroseovarius sp. Z3 | A. crassostreae CV919-312T | A. halocynthiae MA1-10T | A. marinus A6024T | A. pelagivivens GYSW-22T | A. sediminilitoris DSM 29439T | |
|---|---|---|---|---|---|---|
| Aliiroseovarius sp. Z3 | * | 84.51% | 84.33% | 84.87% | 84.49% | 84.95% |
| A. crassostreae CV919-312T | 84.51% | * | 84.82% | 84.95% | 85.04% | 83.92% |
| A. halocynthiae MA1-10T | 84.33% | 84.81% | * | 85.82% | 85.52% | 83.69% |
| A. marinus A6024T | 84.87% | 84.94% | 85.83% | * | 86.50% | 83.62% |
| A. pelagivivens GYSW-22T | 84.51% | 85.02% | 85.52% | 86.50% | * | 83.63% |
| A. sediminilitoris DSM 29439T | 84.95% | 83.92% | 83.69% | 83.61% | 83.65% | * |
Tab. 3
Genome features summary of Z3 and other five similar type strains from Aliiroseovarius genus"
| 名称 | 大小/Mb | GC含量/% | 编码蛋白 | 分离来源 |
|---|---|---|---|---|
| Aliiroseovarius sp. Z3 | 3.53 | 59.4 | 3509 | 中国浙江宁波: 凡纳滨对虾肠道 |
| A. crassostreae CV919-312T | 3.73 | 58.4 | 3693 | 美国缅因州达马里斯科塔河: 患病牡蛎 |
| A. sediminilitoris DSM 29439T | 3.41 | 58.7 | 3323 | 中国南部吉野岛: 近海沉积物 |
| A. marinus A6024T | 3.13 | 59.9 | 3027 | 中国山东日照: 海水 |
| A. halocynthiae MA1-10T | 3.39 | 57.1 | 3349 | 南海: 海鞘 |
| A. pelagivivens GYSW-22T | 3.33 | 58.1 | 3218 | 韩国巨济岛: 海水 |

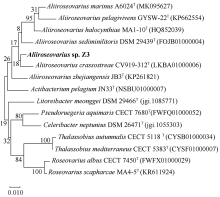
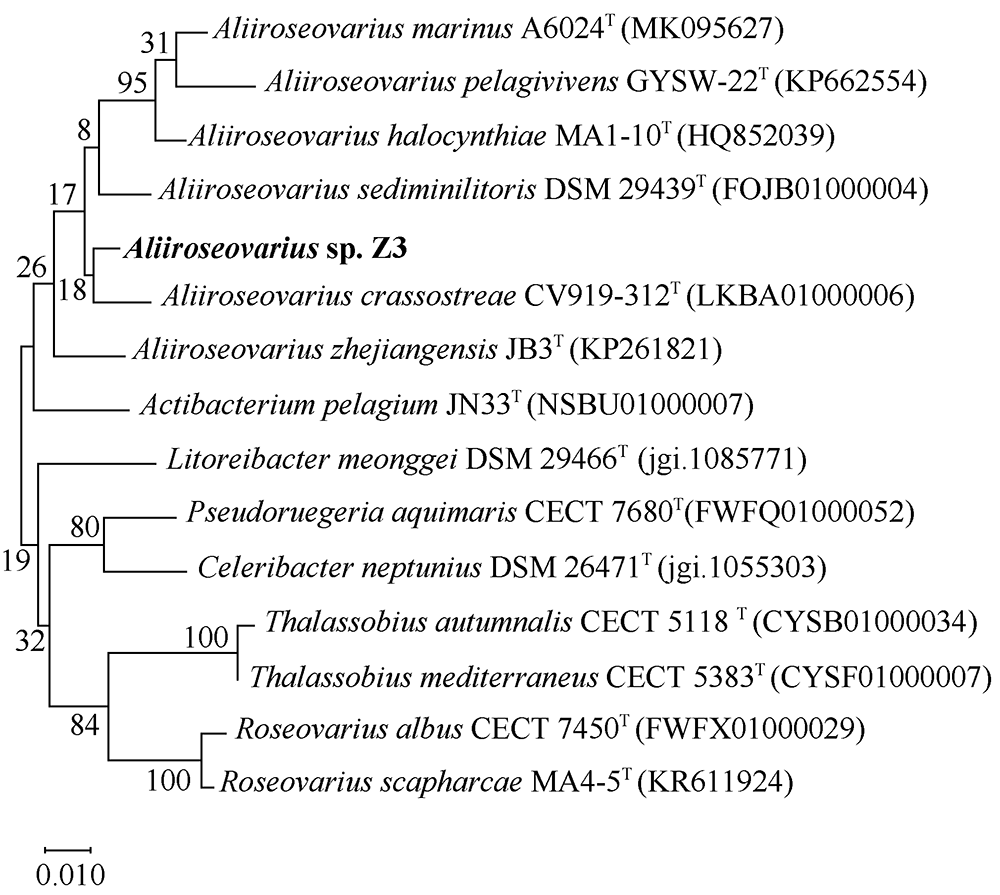
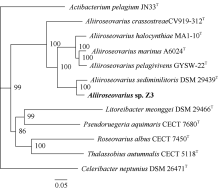
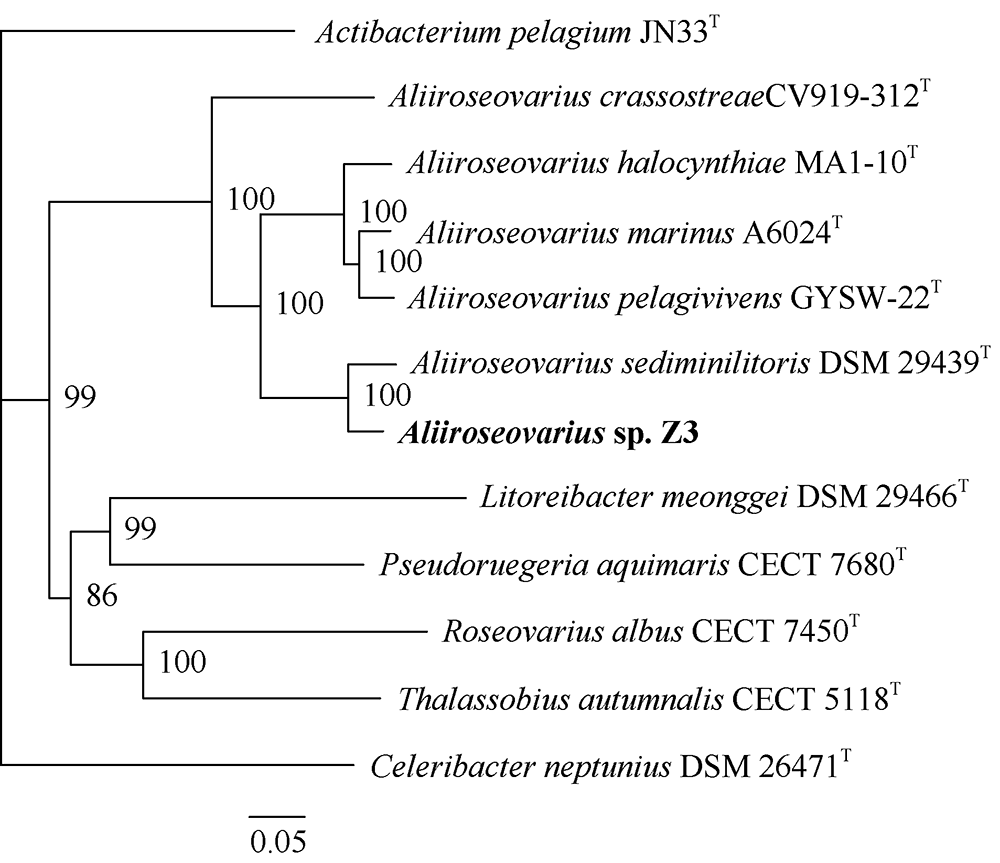
Fig. 5
Synteny blocks between Aliiroseovarius sp. Z3 and other five Aliiroseovarius spp., Aliiroseovarius. crassostreae CV919-312T (a), Aliiroseovarius sediminilitoris DSM 29439T (b), Aliiroseovarius. halocynthiae MA1-10T (c), Aliiroseovarius marinus A6024T (d), and Aliiroseovarius pelagivivens GYSW-22T (e). The value in the figure indicates the size of the gene fragment"

Tab. 4
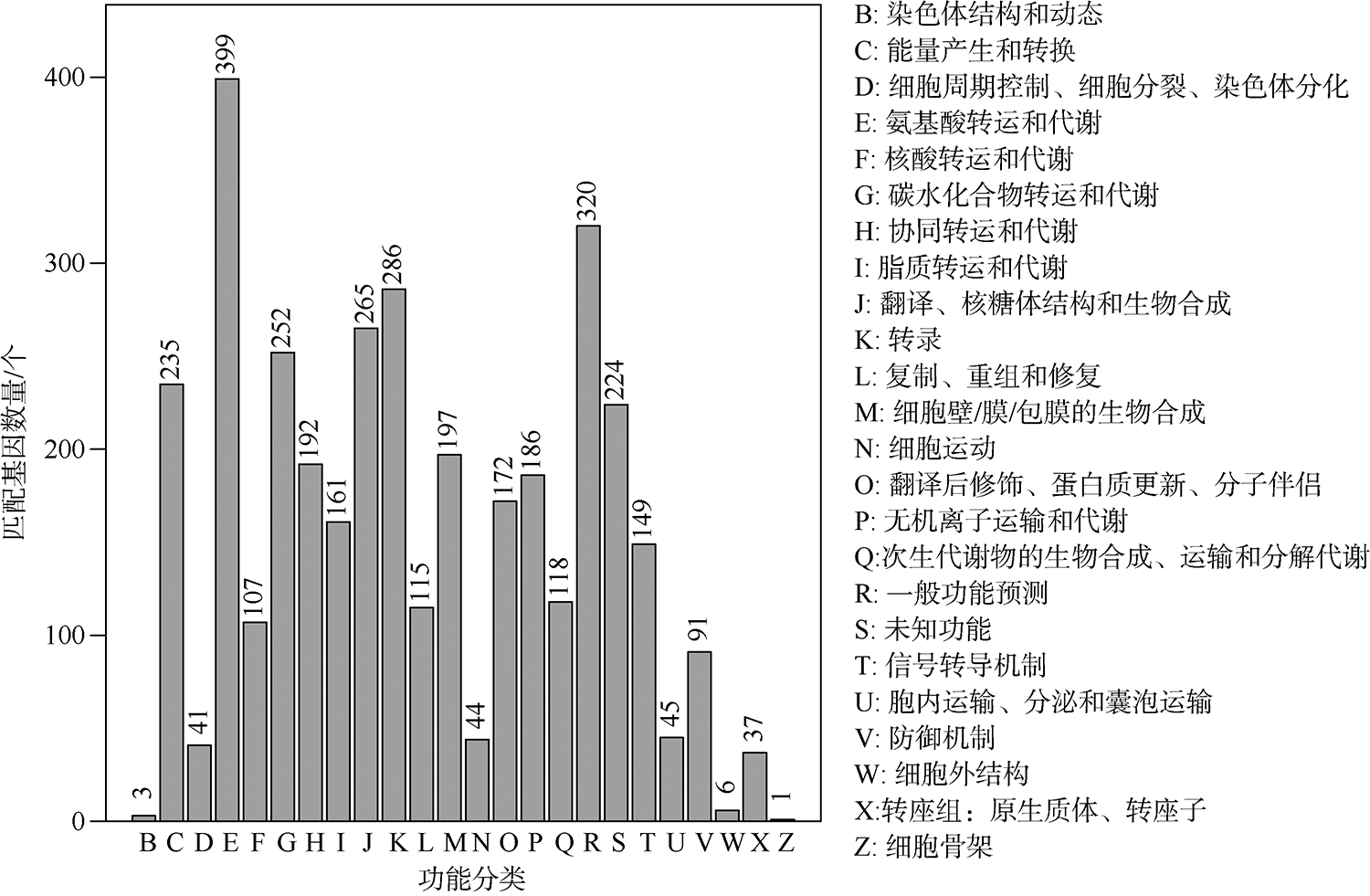
The number of core orthologous gene clusters of six Aliiroseovarius spp. and specific 418 genes in the genome of Aliiroseovarius sp. Z3 based on COG categories analysis"
| COG分类 | 核心基因/个 | Z3的特异性基因/个 |
|---|---|---|
| 未分类(Unassigned) | 992 | 220 |
| 能量产生与转换(C) | 149 | 13 |
| 细胞周期调控、细胞分裂、染色体分区(D) | 9 | 2 |
| 氨基酸转运与代谢(E) | 305 | 14 |
| 核苷酸转运与代谢(F) | 22 | 2 |
| 碳水化合物的转运与代谢(G) | 139 | 15 |
| 辅酶转运与代谢(H) | 97 | 6 |
| 脂类转运与代谢(I) | 78 | 6 |
| 翻译、核糖体结构和生物合成(J) | 30 | 3 |
| 转录(K) | 217 | 12 |
| 复制、重组和修复(L) | 31 | 12 |
| 细胞壁/膜/包体生物合成(M) | 76 | 10 |
| 细胞运动(N) | 2 | 0 |
| 翻译后修饰、蛋白质周转和分子伴侣(O) | 46 | 8 |
| 无机离子转运与代谢(P) | 235 | 12 |
| 次生代谢物的生物合成、转运和分解代谢(Q) | 48 | 2 |
| 功能未知(S) | 184 | 68 |
| 信号转导机制(T) | 110 | 4 |
| 细胞内运输、分泌和囊泡运输(U) | 17 | 3 |
| 防御机制(V) | 44 | 6 |
| [1] | 刘志迎, 许海, 詹旭, 等, 2019. 蓝藻水华对太湖水柱反硝化作用的影响[J]. 环境科学, 40(3): 1261-1269. |
| LIU ZHIYING, XU HAI, ZHJAN XU, et al, 2019. Influence of Cyanobacterial blooms on denitrification rate in shallow Lake Taihu, China[J]. Environmental Science, 40(3): 1261-1269. (in Chinese with English abstract) | |
| [2] |
ABDELHAMED H, NHO S W, KARSI A, et al, 2021. The role of denitrification genes in anaerobic growth and virulence of Flavobacterium columnare[J]. Journal of Applied Microbiology, 130(4): 1062-1074.
doi: 10.1111/jam.v130.4 |
| [3] |
BOETTCHER K J, BARBER B J, SINGER J T, 1999. Use of antibacterial agents to elucidate the etiology of juvenile oyster disease (JOD) in Crassostrea virginica and numerical dominance of an α-proteobacterium in JOD-affected animals[J]. Applied and Environmental Microbiology, 65(6): 2534-2539.
doi: 10.1128/AEM.65.6.2534-2539.1999 |
| [4] |
BOETTCHER K J, GEAGHAN K K, MALOY A P, et al, 2005. Roseovarius crassostreae sp. nov., a member of the Roseobacter clade and the apparent cause of juvenile oyster disease (JOD) in cultured Eastern oysters[J]. International Journal of Systematic and Evolutionary Microbiology, 55(4): 1531-1537.
doi: 10.1099/ijs.0.63620-0 |
| [5] |
BUCHAN A, GONZALEZ J M, MORAN M A, 2005. Overview of the marine Roseobacter lineage[J]. Applied and Environmental Microbiology, 71(10): 5665-5677.
doi: 10.1128/AEM.71.10.5665-5677.2005 |
| [6] |
CAMARGO J A, ALONSO A, SALAMANCA A, 2005. Nitrate toxicity to aquatic animals: a review with new data for freshwater invertebrates[J]. Chemosphere, 58(9): 1255-1267.
doi: 10.1016/j.chemosphere.2004.10.044 |
| [7] |
CASTRESANA J, 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis[J]. Molecular Biology and Evolution, 17(4): 540-552.
doi: 10.1093/oxfordjournals.molbev.a026334 |
| [8] |
CHOI D H, CHO B C, 2006. Citreimonas salinaria gen. nov., sp. nov., a member of the Roseobacter clade isolated from a solar saltern[J]. International Journal of Systematic and Evolutionary Microbiology, 56(12): 2799-2803.
doi: 10.1099/ijs.0.64373-0 |
| [9] |
DARLING A C E, MAU B, BLATTNER F R, et al, 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements[J]. Genome Research, 14(7): 1394-1403.
doi: 10.1101/gr.2289704 |
| [10] |
EDGAR R C, 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput[J]. Nucleic Acids Research, 32(5): 1792-1797.
doi: 10.1093/nar/gkh340 |
| [11] |
EMMS D M, KELLY S, 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics[J]. Genome Biology, 20(1): 238.
doi: 10.1186/s13059-019-1832-y |
| [12] |
GRAHAM D W, TRIPPETT C, DODDS W K, et al, 2010. Correlations between in situ denitrification activity and nir-gene abundances in pristine and impacted prairie streams[J]. Environmental Pollution, 158(10): 3225-3229.
doi: 10.1016/j.envpol.2010.07.010 |
| [13] |
GUO HAIPENG, HUANG LEI, HU SONGTAO, et al, 2020. Effects of carbon/nitrogen ratio on growth, intestinal microbiota and metabolome of shrimp (Litopenaeus vannamei)[J]. Frontiers in Microbiology, 11: 652.
doi: 10.3389/fmicb.2020.00652 |
| [14] |
HUANG LEI, GUO HAIPENG, et al, 2020. The bacteria from large-sized bioflocs are more associated with the shrimp gut microbiota in culture system[J]. Aquaculture, 523: 735159.
doi: 10.1016/j.aquaculture.2020.735159 |
| [15] |
INGLIN R C, MEILE L, STEVENS M J A, 2018. Clustering of pan- and core-genome of Lactobacillus provides novel evolutionary insights for differentiation[J]. BMC Genomics, 19(1): 284.
doi: 10.1186/s12864-018-4601-5 |
| [16] |
JUNG Y T, LEE J S, OH K H, et al, 2011. Roseovarius marinus sp. nov., isolated from seawater[J]. International Journal of Systematic and Evolutionary Microbiology, 61(2): 427-432.
doi: 10.1099/ijs.0.019828-0 |
| [17] |
KANEHISA M, GOTO S, 2000. KEGG: kyoto encyclopedia of genes and genomes[J]. Nucleic Acids Research, 28(1): 27-30.
doi: 10.1093/nar/28.1.27 |
| [18] |
KENT A G, GARCIA C A, MARTINY A C, 2018. Increased biofilm formation due to high-temperature adaptation in marine Roseobacter[J]. Nature Microbiology, 3(9): 989-995.
doi: 10.1038/s41564-018-0213-8 |
| [19] | KESSNER L, 2015. Characterization of biofilm formation, chemotaxis, and the genome of Aliiroseovarius crassostreae[D]. Kingston: University of Rhode Island. |
| [20] |
KIM M, OH H S, PARK S C, et al, 2014. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes[J]. International Journal of Systematic and Evolutionary Microbiology, 64(Pt 2): 346-351.
doi: 10.1099/ijs.0.059774-0 |
| [21] |
KIM Y O, KONG H J, Park S, et al, 2012. Roseovarius halocynthiae sp. nov., isolated from the sea squirt Halocynthia roretzi[J]. International Journal of Systematic and Evolutionary Microbiology, 62(Pt 4): 931-936.
doi: 10.1099/ijs.0.031674-0 |
| [22] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Molecular Biology and Evolution, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 |
| [23] |
LEE I, KIM Y O, PARK S C, et al, 2016. OrthoANI: An improved algorithm and software for calculating average nucleotide identity[J]. International Journal of Systematic and Evolutionary Microbiology, 66(2): 1100-1103.
doi: 10.1099/ijsem.0.000760 |
| [24] |
LI WEIZHONG, GODZIK A, 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences[J]. Bioinformatics, 22(13): 1658-1659.
doi: 10.1093/bioinformatics/btl158 |
| [25] |
LUO HAIWEI, LÖYTYNOJA A, MORAN M A, 2012. Genome content of uncultivated marine Roseobacters in the surface ocean[J]. Environmental Microbiology, 14(1): 41-51.
doi: 10.1111/j.1462-2920.2011.02528.x |
| [26] |
MASELLA A P, BARTRAM A K, TRUSZKOWSKI J M, et al, 2012. PANDAseq: paired-end assembler for illumina sequences[J]. BMC Bioinformatics, 13: 31.
doi: 10.1186/1471-2105-13-31 |
| [27] |
MEIER-KOLTHOFF J P, AUCH A F, KLENK H P, et al, 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions[J]. BMC Bioinformatics, 14(1): 60.
doi: 10.1186/1471-2105-14-60 |
| [28] |
MOU XIAOZHEN, SUN SHULEI, EDWARDS R A, et al, 2008. Bacterial carbon processing by generalist species in the coastal ocean[J]. Nature, 451(7179): 708-711.
doi: 10.1038/nature06513 |
| [29] | NATALE D A, SHANKAVARAM U T, GALPERIN M Y, et al, 2000. Towards understanding the first genome of a crenarchaeon by genome annotation using clusters of orthologous groups of proteins (COGs)[J]. Genome Biology, 1(5): RESEARCH0009. |
| [30] |
PARK S, PARK J M, KANG C H, et al, 2015. Aliiroseovarius pelagivivens gen. nov., sp. nov., isolated from seawater, and reclassification of three species of the genus Roseovarius as Aliiroseovarius crassostreae comb. nov., Aliiroseovarius halocynthiae comb. nov. and Aliiroseovarius sediminilitoris comb. nov[J]. International Journal of Systematic and Evolutionary Microbiology, 65(Pt 8): 2646-2652.
doi: 10.1099/ijs.0.000315 |
| [31] |
PARK S, YOON J H, 2013. Roseovarius sediminilitoris sp. nov., isolated from seashore sediment[J]. International Journal of Systematic and Evolutionary Microbiology, 63(5): 1741-1745.
doi: 10.1099/ijs.0.043737-0 |
| [32] |
PHILIPPOT L, 2002. Denitrifying genes in bacterial and Archaeal genomes[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1577(3): 355-376.
doi: 10.1016/S0167-4781(02)00420-7 |
| [33] | PUJALTE M J, LUCENA T, RUVIRA M A, et al, 2014. The family Rhodobacteraceae[M]// The Prokaryotes. Berlin, Heidelberg: Springer: 439-512. |
| [34] | REISCH C R, MORAN M A, WHITMAN W B, 2011. Bacterial catabolism of dimethylsulfoniopropionate (DMSP)[J]. Frontiers in Microbiology, 2: 172. |
| [35] | RICHTER M, ROSSELLÓ-MÓRA R, 2009. Shifting the genomic gold standard for the prokaryotic species definition[J]. Proceedings of the National Academy of Sciences of the United States of America, 106(45): 19126-19131. |
| [36] | SHARPE G C, GIFFORD S M, SEPTER A N, 2020. A model Roseobacter, Ruegeria pomeroyi DSS-3, Employs a diffusible killing mechanism to eliminate competitors[J]. mSystems, 5(4): e00443-20. |
| [37] |
STAMATAKIS A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies[J]. Bioinformatics, 30(9): 1312-1313.
doi: 10.1093/bioinformatics/btu033 |
| [38] |
SUN SHENGMING, GE XIANPING, ZHU JIAN, et al, 2014. Identification and mRNA expression of antioxidant enzyme genes associated with the oxidative stress response in the Wuchang bream (Megalobrama amblycephala Yih) in response to acute nitrite exposure[J]. Comparative Biochemistry and Physiology Part C: Toxicology and Pharmacology, 159: 69-77.
doi: 10.1016/j.cbpc.2013.09.005 |
| [39] |
XING WEI, LI DESHENG, LI JINLONG, et al, 2016. Nitrate removal and microbial analysis by combined micro- electrolysis and autotrophic denitrification[J]. Bioresource Technology, 211: 240-247.
doi: 10.1016/j.biortech.2016.03.044 |
| [40] |
YU ZEHUI, GENG YI, WANG KAIYU, et al, 2017. Complete genome sequence of Vibrio mimicus strain SCCF01 with potential application in fish vaccine development[J]. Virulence, 8(6): 1028-1030.
doi: 10.1080/21505594.2016.1250996 |
| [41] |
ZEB S, GULFAM S M, BOKHARI H, 2020. Comparative core/pan genome analysis of Vibrio cholerae isolates from Pakistan[J]. Infection, Genetics and Evolution, 82: 104316.
doi: 10.1016/j.meegid.2020.104316 |
| [42] |
ZHANG XIAN, LIU ZHENGHUA, WEI GUANYUN, et al, 2018. In silico genome-wide analysis reveals the potential links between core genome of Acidithiobacillus thiooxidans and its autotrophic lifestyle[J]. Frontiers in Microbiology, 9: 1255.
doi: 10.3389/fmicb.2018.01255 |
| [1] | MO Danyang, NING Zhiming, YANG Bin, XIA Ronglin, LIU Zhijin. Response of dissimilatory nitrate reduction processes in coral reef sediments of the Weizhou island to temperature changes [J]. Journal of Tropical Oceanography, 2024, 43(4): 137-143. |
| [2] | HUO Jiaxin, LI Yingxin, SONG Yan, ZHU Qing, ZHOU Weihua, YUAN Xiangcheng, HUANG Hui, LIU Sheng. Complete mitochondrial genome of Cladopsammia gracilis and Rhizopsammia wettsteini (Scleractinia, Dendrophylliidae) and its phylogenetic implications* [J]. Journal of Tropical Oceanography, 2024, 43(3): 22-30. |
| [3] | LI Nenghui, HUANG Qing, LI Hang, ZENG Jun, WU Kefeng, TAN Huaqiang. Taxonomic study of four species of Gracilaria (Gracilariaceae, Rhodophyta) in Zhanjiang based on morphological and molecular data [J]. Journal of Tropical Oceanography, 2024, 43(2): 34-47. |
| [4] | HONG Yiguo, JIAO Lijing, WU Jiapeng, LONG Aimin, WANG Wei. Progress on the community distribution and ecological functions of nitrite-oxidizing bacteria [J]. Journal of Tropical Oceanography, 2021, 40(2): 139-146. |
|
||