Journal of Tropical Oceanography ›› 2023, Vol. 42 ›› Issue (6): 89-100.doi: 10.11978/2023036CSTR: 32234.14.2023036
• Marine Biology • Previous Articles Next Articles
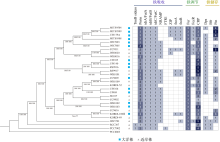
Adaptive mechanisms of iron limitation on the marine Synechococcus based on comparative genomics
MU Rong( ), ZHU Zhu(
), ZHU Zhu( ), ZHANG Ruifeng
), ZHANG Ruifeng
- School of Oceanography, Shanghai Jiao Tong University, Shanghai 200030, China
-
Received:2023-03-23Revised:2023-04-28Online:2023-11-10Published:2023-11-28 -
Supported by:National Natural Science Foundation of China(41890801); International Cooperation in Science and Technology Innovation between Governments(2022YFE0136500)
Cite this article
MU Rong, ZHU Zhu, ZHANG Ruifeng. Adaptive mechanisms of iron limitation on the marine Synechococcus based on comparative genomics[J].Journal of Tropical Oceanography, 2023, 42(6): 89-100.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Functional genes related to iron limitation"
| 基因分类 | 基因 | 基因描述 | KEGG 编号 | |
|---|---|---|---|---|
| 铁吸收 | 铁载体相关基因 | TonB sidero | TonB dependent receptor | K02014 |
| 孔蛋白基因 | Porin | Outer membrane protein insertion porin family | K07277 | |
| Fe3+转运基因 | idiA/FutA | Iron deficiency induced protein A / iron(Ⅲ) transport system substrate-binding protein | K02012 | |
| idiB/FutB | ABC-type Fe3+ transport system permease component / iron(Ⅲ) transport system permease protein | K02011 | ||
| idiC/FutC | ATP binding component / iron(Ⅲ) transport system ATP-binding protein | K02010 | ||
| 二价金属转运基因 | NRAMP | Natural resistance associated macrophage protein, Nramp | K03322 | |
| FTR1 | FTR1 family iron permease | K07243 | ||
| ZIP | ZRT / IRT-like Protein | K14709 | ||
| Fe2+转运基因 | FeoA | Ferrous iron transport protein A | K04758 | |
| FeoB | Ferrous iron transport protein B | K04759 | ||
| 铁稳态 | — | Fur | Ferric uptake regulator | K03711 |
| FecR | Periplasmic ferric-dicitrate binding protein FerR | K07165 | ||
| CRP | Cyclic AMP receptor protein | K10914 | ||
| 铁储存 | 铁储存蛋白 | Dps | DNA-binding stress protein | K04047 |
| Bfr | Bacterioferritin | K03594 | ||
| Ftn | Ferritins | K02217 | ||
Tab. 2
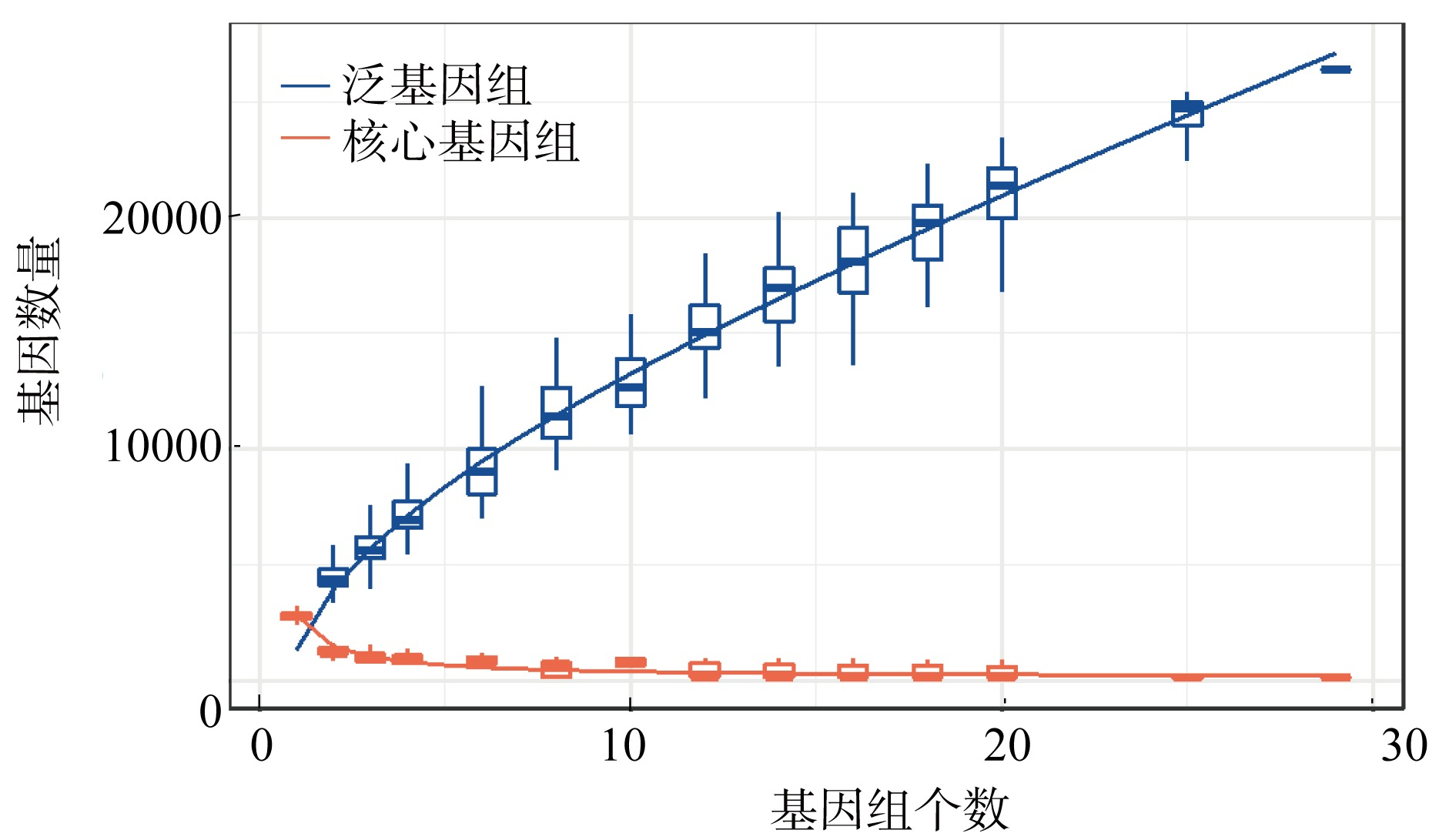
Genome features of the 29 analyzed marine Synechococcus strains"
| 分离地点 | 藻株 | 基因组编号 | 类型 | 基因组大小/bp | GC含量/% | 完整度/% | 污染度/% | |
|---|---|---|---|---|---|---|---|---|
| 大西洋马尾藻海 | WH8102 | GCA_000195975.1 | 大洋 | 2434428 | 59.4 | 99.46 | 0 | |
| WH7803 | GCA_000063505.1 | 大洋 | 2366980 | 60.2 | 99.18 | 0 | ||
| WH8103 | GCA_001182765.1 | 大洋 | 2429688 | 59.5 | 99.73 | 0.27 | ||
| UW69 | GCA_900474185.1 | 大洋 | 2372121 | 58.7 | 100 | 0 | ||
| UW140 | GCA_900474295.1 | 大洋 | 2704142 | 57.8 | 99.73 | 0 | ||
| UW105 | GCA_900473935.1 | 大洋 | 2659417 | 57.5 | 99.46 | 0 | ||
| UW106 | GCA_900474015.1 | 大洋 | 2479535 | 58 | 99.73 | 0.27 | ||
| WH8109 | GCA_000161795.2 | 大洋 | 2111515 | 60.1 | 99.46 | 0 | ||
| 赤道太平洋 | UW179A | GCA_900473965.1 | 大洋 | 3057835 | 54.5 | 100 | 0.41 | |
| MITS9508 | GCA_001632165.1 | 大洋 | 2502434 | 56.1 | 100 | 0.27 | ||
| MITS9509 | GCA_001631935.1 | 大洋 | 3087928 | 55.4 | 99.18 | 0.82 | ||
| MITS9504 | GCA_001632105.1 | 大洋 | 3087293 | 55.4 | 99.73 | 1.09 | ||
| 东太平洋加利福尼亚海流 | CC9605 | GCA_000012625.1 | 近岸 | 2510659 | 59.2 | 99.73 | 0 | |
| CC9311 | GCA_000014585.1 | 近岸 | 2606748 | 52.5 | 99.73 | 0 | ||
| CC9902 | GCA_000012505.1 | 近岸 | 2234828 | 54.2 | 99.46 | 0 | ||
| 中国东海 | KORDI-52 | GCA_000737595.1 | 近岸 | 2572069 | 59.1 | 100 | 0 | |
| KORDI-49 | GCA_000737575.1 | 近岸 | 2585813 | 61.4 | 99.37 | 0 | ||
| 地中海 | RCC307 | GCA_000063525.1 | 近岸 | 2224914 | 60.8 | 99.64 | 0 | |
| BL107 | GCA_000153805.1 | 近岸 | 2285034 | 54.2 | 99.46 | 0 | ||
| 红海亚喀巴湾 | RS9916 | GCA_000153825.1 | 近岸 | 2664873 | 59.8 | 99.73 | 0 | |
| RS9917 | GCA_000153065.1 | 近岸 | 2584918 | 64.5 | 99.46 | 0 | ||
| 波多黎马格耶斯岛 | PCC7002 | GCA_000019485 | 近岸 | 3409935 | 49.2 | 100 | 0 | |
| 西太平洋 | KORDI-100 | GCA_000737535.1 | 大洋 | 2789000 | 57.5 | 99.46 | 0 | |
| 新英格兰大陆架 | WH8020 | GCA_001040845.1 | 近岸 | 2661166 | 53.1 | 99.73 | 0.27 | |
| 伍兹霍尔沿海海水 | WH8101 | GCA_004209775.1 | 近岸 | 2630292 | 63.3 | 100 | 0.82 | |
| 北大西洋潮间带 | WH8016 | GCA_000230675.2 | 近岸 | 2694843 | 54.1 | 99.18 | 0 | |
| 北大西洋 | WH7805 | GCA_000153285.1 | 大洋 | 2627046 | 57.6 | 99.73 | 0 | |
| 北大西洋长岛海峡 | WH5701 | GCA_000153045 | 近岸 | 3280236 | 65.4 | 99.18 | 7.84 | |
| 东太平洋 | CC9616 | GCA_000515235.1 | 大洋 | 2645910 | 56.6 | 99.46 | 0.27 | |
Tab. 3
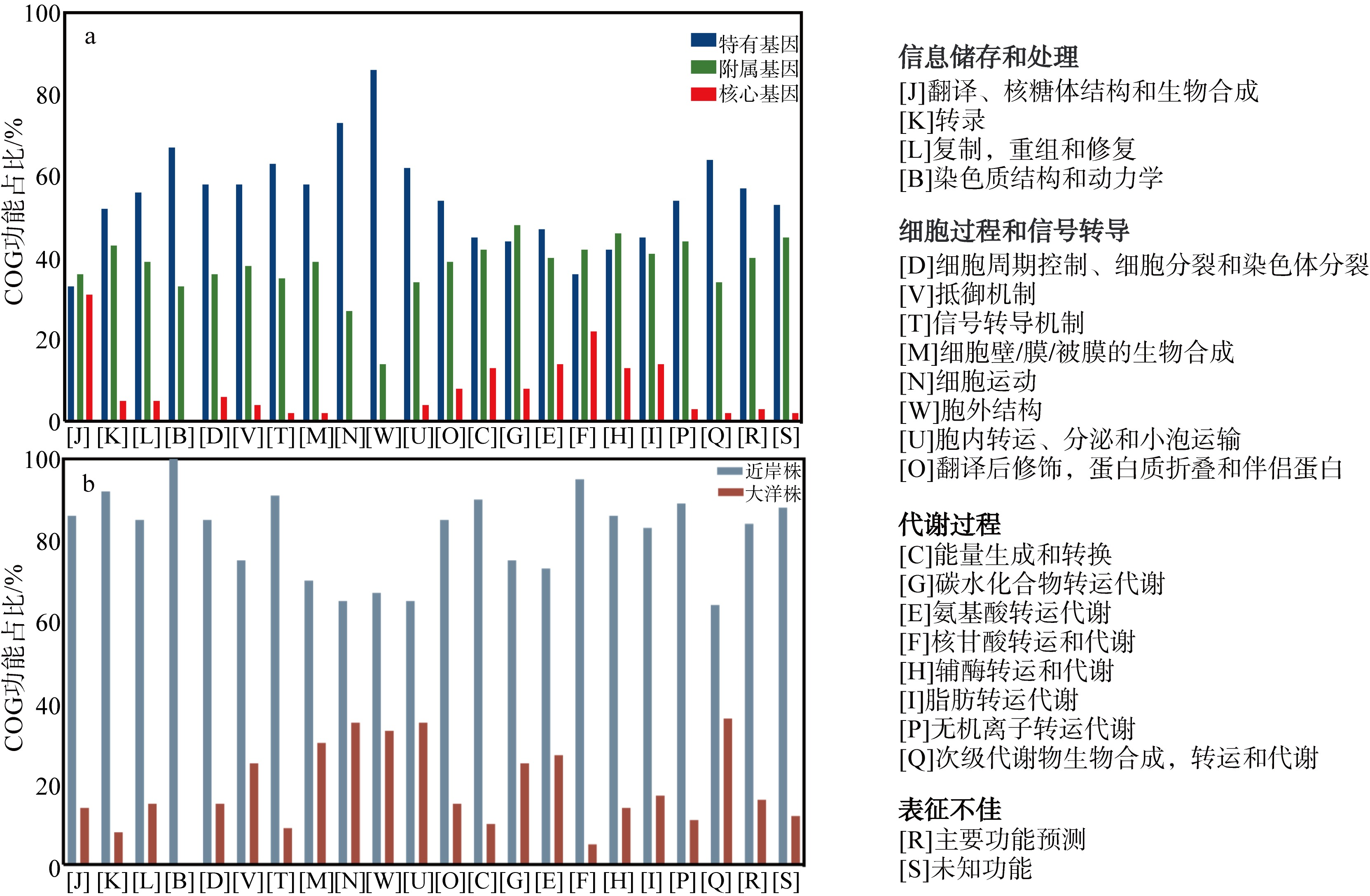
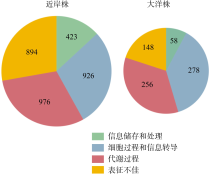
Differences in adaptive mechanisms for iron limitation of coastal and oceanic Synechococcus"
| 相关基因/蛋白 | 海洋聚球藻 | ||
|---|---|---|---|
| 近岸株 | 大洋株 | ||
| 铁吸收 | 铁载体 | 合成和利用铁载体的能力较强 | 合成和利用铁载体的能力较弱 |
| 孔蛋白 | 存在 | 存在 | |
| Fe(Ⅲ)转运蛋白 | 存在, 基因表达量相对较低 | 存在, 基因表达量相对较高 | |
| Fe(Ⅱ)特异性转运蛋白 | 普遍存在 | 除生态型CRD1中不存在 | |
| 铁稳态 | Fur家族(FurA、Zur、PerR) | 存在, 具有更复杂的调节能力 | 存在 |
| FecR | 存在 | 仅在生态型CRD1中广泛分布 | |
| CRP | 存在 | 存在 | |
| 铁储存 | Ftn | 拷贝数较多 | 仅在生态型CRD1中拥有多拷贝数 |
| Bfr | 存在 | 不存在 | |
| Dps | 存在 | 存在 | |
| [1] |
彭铭烨, 黄婷, 张晓娟, 等, 2023. 比较基因组学解析谷物醋醋醅中巴氏醋杆菌和欧洲驹形杆菌的功能差异[J]. 微生物学报, 63(2): 638-655.
|
|
|
|
| [2] |
郑强, 贺博闻, 史文卿, 等, 2023. 海洋超微型蓝细菌聚球藻的生态学研究进展[J]. 厦门大学学报(自然科学版), 62(3): 301-313.
|
|
|
|
| [3] |
doi: 10.1111/emi.v22.5 |
| [4] |
pmid: 12829269 |
| [5] |
pmid: 17804072 |
| [6] |
doi: 10.1111/fem.1996.21.issue-2 |
| [7] |
doi: 10.1093/bioinformatics/btz848 |
| [8] |
doi: 10.1038/s41396-020-00775-z |
| [9] |
|
| [10] |
doi: 10.7717/peerj.1522 |
| [11] |
doi: 10.3389/fmicb.2020.567431 |
| [12] |
doi: 10.1128/msystems.00656-22 |
| [13] |
doi: 10.1186/gb-2008-9-5-r90 |
| [14] |
doi: 10.1111/emi.2014.16.issue-3 |
| [15] |
|
| [16] |
doi: 10.1128/AEM.69.5.2430-2443.2003 |
| [17] |
doi: 10.3389/fmicb.2020.00165 pmid: 32184761 |
| [18] |
doi: 10.1111/j.1462-2920.2011.02539.x pmid: 21883791 |
| [19] |
doi: 10.1007/s10534-009-9235-2 pmid: 19343508 |
| [20] |
|
| [21] |
doi: 10.1038/nmeth.4285 pmid: 28481363 |
| [22] |
doi: 10.1093/molbev/msw009 pmid: 26769030 |
| [23] |
doi: 10.1128/JB.183.9.2779-2784.2001 |
| [24] |
|
| [25] |
doi: 10.1093/bioinformatics/btz188 pmid: 30865266 |
| [26] |
doi: 10.3390/life5010841 |
| [27] |
doi: 10.1016/S0304-4203(01)00074-3 |
| [28] |
doi: 10.1016/j.jmb.2007.09.010 |
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
doi: 10.1128/MMBR.00012-07 pmid: 17804665 |
| [34] |
|
| [35] |
doi: 10.4319/lo.2008.53.1.0400 |
| [36] |
pmid: 12738853 |
| [37] |
doi: 10.3389/fmicb.2012.00043 pmid: 22408637 |
| [38] |
doi: 10.1093/molbev/msu300 |
| [39] |
doi: 10.1093/bioinformatics/btv421 pmid: 26198102 |
| [40] |
|
| [41] |
doi: 10.1093/nar/gkab776 |
| [42] |
doi: 10.1038/nbt.4229 pmid: 30148503 |
| [43] |
doi: 10.1101/gr.186072.114 pmid: 25977477 |
| [44] |
doi: 10.1038/ismej.2013.228 |
| [45] |
doi: 10.1016/j.tim.2021.06.001 |
| [46] |
doi: 10.1111/emi.v23.1 |
| [47] |
doi: 10.1023/A:1006282714942 |
| [48] |
doi: 10.1111/emi.2009.11.issue-2 |
| [49] |
doi: 10.1128/AEM.68.3.1180-1191.2002 |
| [50] |
doi: 10.1111/emi.v22.11 |
| [51] |
doi: 10.1007/s10872-017-0457-6 |
| [52] |
|
| [53] |
doi: 10.1128/MMBR.00035-08 pmid: 19487728 |
| [54] |
doi: 10.1016/0304-4203(95)00035-P |
| [55] |
|
| [56] |
doi: 10.1038/nature21058 |
| [57] |
doi: 10.1111/1462-2920.14876 pmid: 31769166 |
| [58] |
doi: 10.1038/s41579-019-0294-2 pmid: 31792365 |
| [59] |
doi: 10.1128/AEM.67.12.5444-5452.2001 |
| [60] |
doi: 10.4319/lo.1999.44.4.1002 |
| [61] |
doi: 10.1128/msystems.01522-21 |
| [62] |
|
| [63] |
doi: 10.1007/s00248-017-1021-z |
| [1] | MAO Yingjin, GAO Beile. Comparative genomic analysis of the distribution and evolution of quorum sensing pathways in the Vibrio genus [J]. Journal of Tropical Oceanography, 2022, 41(1): 28-41. |
|
||