Journal of Tropical Oceanography ›› 2021, Vol. 40 ›› Issue (2): 27-38.doi: 10.11978/2020055CSTR: 32234.14.2020055
• Marine Biology • Previous Articles Next Articles
Functional study of coupling protein CheV and CZB domain of chemoreceptors in the Epsilon-proteobacteria chemotaxis signaling pathway
LIU Yugeng1,2( ), MAO Yingjin1,2, ZHANG Canchuan1,2, GAO Beile1(
), MAO Yingjin1,2, ZHANG Canchuan1,2, GAO Beile1( )
)
- 1. CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, Guangdong Key Laboratory of Marine Materia Medica, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
2. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2020-05-26Revised:2020-06-29Online:2021-03-10Published:2020-07-29 -
Contact:GAO Beile E-mail:yugengliu@163.com;gaob@scsio.ac.cn -
Supported by:National Science Foundation of China(31870064)
CLC Number:
- P735.4
Cite this article
LIU Yugeng, MAO Yingjin, ZHANG Canchuan, GAO Beile. Functional study of coupling protein CheV and CZB domain of chemoreceptors in the Epsilon-proteobacteria chemotaxis signaling pathway[J].Journal of Tropical Oceanography, 2021, 40(2): 27-38.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Strain information"
| 物种名称 | 菌株 | 生物样品序列号 | 生物项目序列号 | 基因组编号 | 纯培养 | 呼吸类型 | 参考文献 |
|---|---|---|---|---|---|---|---|
| Caminibacter mediatlanticus | TB-2 | SAMN10884411 | PRJNA521379 | GCA_006459125.1 | 是 | 厌氧 | |
| Cetia pacifica | TB6 | SAMN03737945 | PRJNA284962 | GCA_003346815.1 | 是 | 厌氧 | |
| Hydrogenimonas sp. | MAG | SAMN00210774 | PRJNA543187 | GCA_005886055.1 | 否 | 兼性厌氧 | |
| Lebetimonas sp.# | JS138 | SAMN03737948 | PRJNA66817 | GCA_003660105.1 | 否 | 厌氧 | |
| Nautilia profundicola | AmH | SAMN03737951 | PRJNA284966 | GCA_003544915.1 | 是 | 厌氧 | |
| Nautilia sp. | PV-1 | SAMN03737952 | PRJNA284967 | GCA_003346755.1 | 是 | 厌氧 | |
| Nitratiruptor sp. | SB155-2 | SAMN03737954 | PRJNA66821 | GCA_003544935.1 | 是 | 兼性厌氧 | |
| Sulfurimonas autotrophica | DSM 16294 | SAMN03737955 | PRJNA66823 | GCA_003346775.1 | 是 | 好氧 | |
| Campylobacter jejuni* | 81-176 | SAMEA4030738 | PRJEB6403 | GCA_900637395.1 | 是 | 微好氧 |
Tab. 3
Campylobacter jejuni 81-176 related genetic information"
| 基因 | 基因编号 | 蛋白结构域示意图 |
|---|---|---|
| cheV | CJJ81176_0311 |  |
| cheW | CJJ81176_0309 |  |
| tlp1 | CJJ81176_1498 |  |
| tlp2 | CJJ81176_0180 |  |
| tlp3# | CJJ81176_1548 |  |
| tlp3# | CJJ81176_1549 |  |
| tlp4 | CJJ81176_0289 |  |
| tlp5* | CJJ81176_0271-4 | Pseudogene |
| tlp6 | CJJ81176_0473 |  |
| tlp7 | CJJ81176_0975 |  |
| tlp8 | CJJ81176_1128 |  |
| tlp9 | CJJ81176_1205 |  |
| tlp10 | CJJ81176_0046 |  |
| aer1 | CJJ81176_1204 |  |
| aer2 | CJJ81176_1206 |  |
Tab.4
Primer sequence"
| 引物名称 | 序列(5’→ 3’) |
|---|---|
| pKNT25+cheV_F | CTCTAGAGGATCCCCGGGTAATGTTTGATGAAAATATCGT |
| pKNT25+cheV_R | GTCATTGAATTCGAGCTCGGTTACCCCTGTTCTTGAGATT |
| pKNT25+cheW_F | CAGGTCGACTCTAGAGGATCCCCGGGTAATGAGTAATGAAAAATTAGAGCAAATTTTGC |
| pKNT25+cheW_R | CTGCATGGTCATTGAATTCGAGCTCGGAAATTCGCGCTTAAGTAAAGCTTCTACTTTG |
| pUT18C+tlp1_F | ACTGCAGGTCGACTCTAGATGAAATTAAAAAGATGCTTTTGGCTT |
| pUT18C+tlp1_R | ATATCGATGAATTGCTCGAGTTAAAATCTTTTTTTACTCACATCTTCAAG |
| pUT18C+tlp4_F | ACGCCACTGCAGGTCGACTCTAGATCTCTCCCCACTTGCAGCTATCCAAACAGGT |
| pUT18C+tlp4_R | TAGTTATATCGATGAATTGCTCGAGTTAAAACCTTTTCTTCTTAACATCTTCTAA |
| pUT18C+tlp6_F | ACGCCACTGCAGGTCGACTCTAGATATGTTTGGAAGTAAAATAAACCATTCTGAT |
| pUT18C+tlp6_R | TAGTTATATCGATGAATTGCTCGAGTTAATGATCTGACTCATCAAGCATTTCTTT |
| pUT18C+tlp7_F | ACTGCAGGTCGACTCTAGATCAATTTATCGAAAAAACTCATAAGGCAGTT |
| pUT18C+tlp7_R | ATATCGATGAATTGCTCGAGTTAAATTTGAAATTGGTTAAGTTCGCTTTC |
| pUT18C+tlp8_F | ACGCCACTGCAGGTCGACTCTAGATATGTTTGGTGCTAAGAAAAATAATACTGAA |
| pUT18C+tlp8_R | TAGTTATATCGATGAATTGCTCGAGTTATGACATCGCTTTAGCAACTTCAGCAGAGCT |
| pUT18C+tlp9_F | ACGCCACTGCAGGTCGACTCTAGATTTAGCAAACATAGAAGTGACAGCAAGATCT |
| pUT18C+tlp9_R | TAGTTATATCGATGAATTGCTCGAGTTATATTTTTAATTTTGCTAAGATTTCAGC |
| pUT18C+tlp10_F | ACTGCAGGTCGACTCTAGATTCAAGTCATCAAAATTCACAAAAACTCAA |
| pUT18C+tlp10_R | ATATCGATGAATTGCTCGAGTTACTGAAAGCTACTTAATTTTTCGGAGAG |
| pUT18C+aer1_F | ACTGCAGGTCGACTCTAGATAAAGAAATAGTTTTGTCTGAAAATGCTTTA |
| pUT18C+aer1_R | ATATCGATGAATTGCTCGAGTTAATTATTTTCTTGTAAGTTAAAAATAAGTTCAT |
| pUT18C+aer2_F | ACTGCAGGTCGACTCTAGATTCAAGAGAAATTTTTTTACAAGAAGATAGT |
| pUT18C+aer2_R | ATATCGATGAATTGCTCGAGTTATTTAGCTTCTTGAAGAGAAAAGATTAGC |
| pUT18C+tlp9 (no CZB-like)_F | ACTGCAGGTCGACTCTAGATTTAGCAAACATAGAAGTGACAGCAAGATCT |
| pUT18C+tlp9 (no CZB-like)_R | ATATCGATGAATTGCTCGAGTTAATAAAGAATATGATCGATTTTAACCAC |
| pCH363+tlp9_F | ACAGCTATGACCATGATTACGCCATTAGCAAACATAGAAGTGACAGCAAGATCT |
| pCH363+tlp9_R | TGGCCTCGCTGGCGGCTGAATTCGATATTTTTAATTTTGCTAAGATTTCAGCACT |
| pGEX-6P+tlp6_F | TCTGTTCCAGGGGCCCCTGGGATCCTTGCTTAGACAACACAAAGATGAGC |
| pGEX-6P+tlp6_R | GCTCGAGTCGACCCGGGAATTCCGGTTATATTTTTAATTTTGCTAAGATTTCAGCAC |
| pGEX-6P+tlp9_F | TCTGTTCCAGGGGCCCCTGGGATCCATGTTTGGAAGTAAAATAAACCATTCTGATCTTC |
| pGEX-6P+tlp9_R | GCTCGAGTCGACCCGGGAATTCCGGTTAATGATCTGACTCATCAAGCATTTC |
| pGEX-6P+tlp9 (no CZB-like)_F | TCTGTTCCAGGGGCCCCTGGGATCCATGTTTGGAAGTAAAATAAACCATTCTGATCTTC |
| pGEX-6P+tlp9 (no CZB-like)_R | GCTCGAGTCGACCCGGGAATTCCGGTTAATCGATTTTAACCACAGACAAAATCAATC |
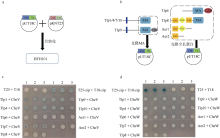
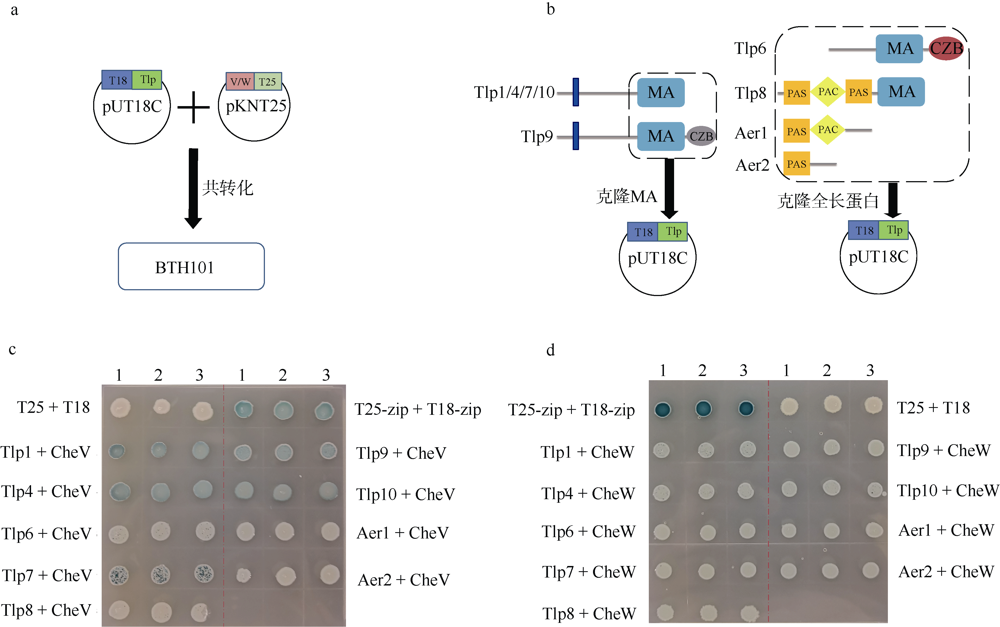
Fig. 2
Using bacterial two-hybrid system to study the interaction between CheV and chemotactic receptors in Campylobacter jejuni 81-176 (a) Schematic diagram of bacterial two-hybrid experiment; (b) Schematic diagram of gene cloning. The left panel shows that for transmembrane Tlps, only MA domains and sequences after the MA domain were cloned into vectors. The right panel shows the full-length sequence of the cytolasmic chemotactic receptor was selected for cloning; (c) Interaction of CheV with Tlps by bacterial two-hybrid experiments; (d) Interaction of CheW with Tlps by bacterial two-hybrid experiments. The shade of color in Figure C and D represents the strength of protein interaction. BTH101 is a strain for bacterial double hybrid system"
Tab. 5
Analysis and statistics of F-Class and CheV in deep sea Epsilon-proteobacteria"
| 菌株 | F类型 | CheV蛋白编号 | 菌株 | F类型 | CheV蛋白编号 |
|---|---|---|---|---|---|
| Campylobacter jejuni* | 1 (F3) | WP_002857365.1 | Nautilia profundicola | 1 (F3) | WP_041361491.1 |
| Caminibacter mediatlanticus | 1 (F3) | WP_007474092.1 | Nautilia sp. PV-1 | 1 (F3) | WP_127679461.1 |
| Cetia pacifica | 1 (F3) | WP_123351600.1 | Nitratiruptor sp. SB155-2 | 1 (F14) | WP_012081979.1 |
| Hydrogenimonas sp. MAG | 1 (F3) | BBG65199.1 | Sulfurimonas autotrophica | 2 (F3/F8) | WP_013326429.1 WP_013327173.1# |
| Lebetimonas sp. JS138 | 1 (F3) | WP_024787070.1 |
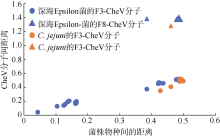
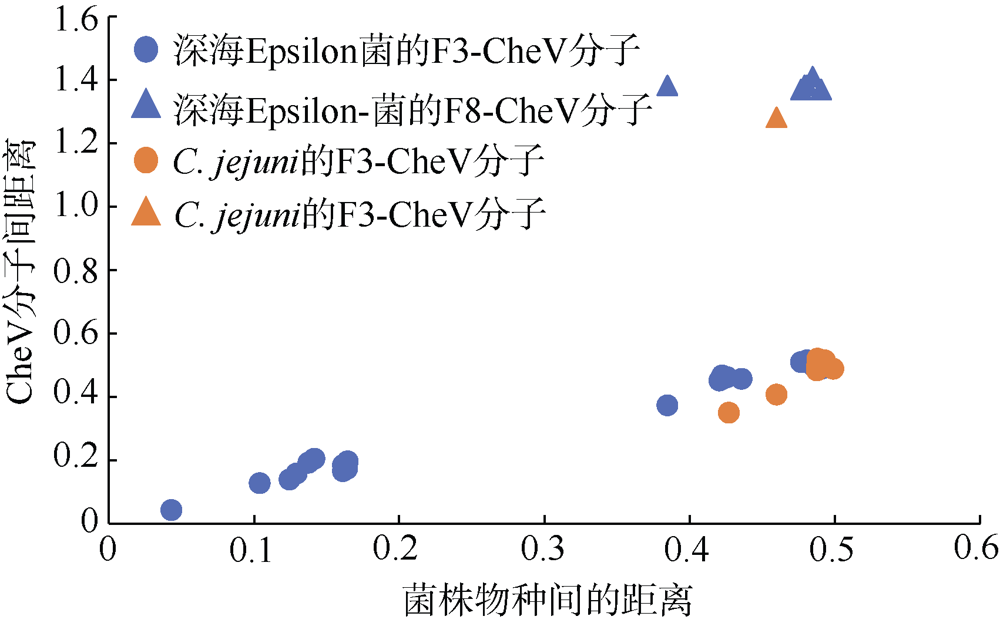
Fig. 3
Pairwise Distances matrix to calculate the evolutionary distance for CheV homologs between deep-sea Epsilon-proteobacteria and Campylobacter jejuni Blue indicates the evolutionary distance between deep-sea Epsilon- proteobacteria. Orange indicates the evolutionary distance between deep-sea Epsilon-proteobacteria and C. jejuni 81-176. Dot shows the distance between F3-CheV in different Epsilon-proteobacteria, and triangle shows the distance between F8-CheV in sulfurimonas autotrophica and F3-CheV in other Epsilon-proteobacteria"
Tab. 6
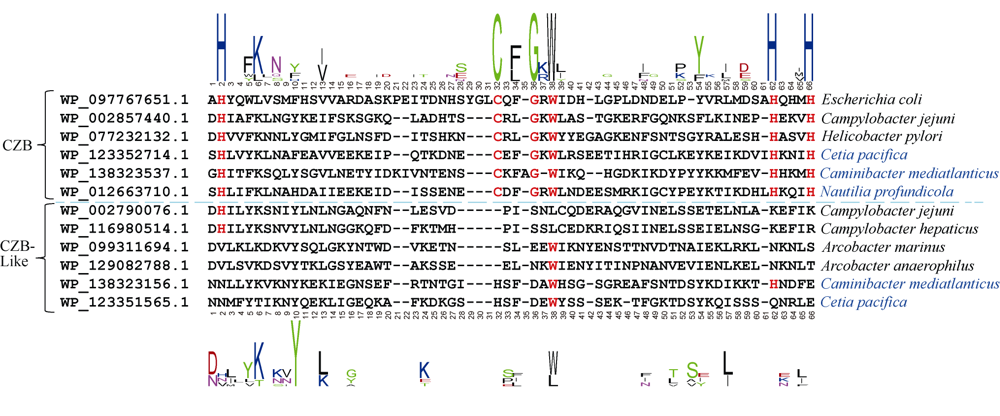
Analysis and statistics of CZB domain in deep sea Epsilon-proteobacteria"
| 菌株 | CZB个数 | 含CZB的蛋白编号 | CZB-like个数 | 含CZB-like的蛋白编号 |
|---|---|---|---|---|
| Campylobacter jejuni* | 1 | WP_002857440.1 | 1 | WP_002790076.1 |
| Caminibacter mediatlanticus | 2 | WP_138323537.1, WP_138323100.1 | 1 | WP_138323156.1 |
| Cetia pacifica | 2 | WP_123352714.1, WP_123352123.1 | 1 | WP_123351565.1 |
| Hydrogenimonas sp. MAG | 5 | BBG65387.1, BBG65955.1, BBG65459.1, BBG65926.1, BBG65424.1 | 0 | 0 |
| Lebetimonas sp. JS138 | 1 | WP_024791915.1 | 0 | 0 |
| Nautilia profundicola | 2 | WP_012663710.1, WP_015902667.1 | 0 | 0 |
| Nautilia sp. PV-1 | 2 | WP_127679641.1, WP_127679872.1 | 0 | 0 |
| Nitratiruptor sp. SB155-2 | 2 | WP_012081746.1, WP_012081977.1 | 0 | 0 |
| Sulfurimonas autotrophica | 3 | WP_013327874.1, WP_041675273.1, WP_013327332.1 | 0 | 0 |
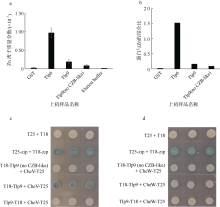
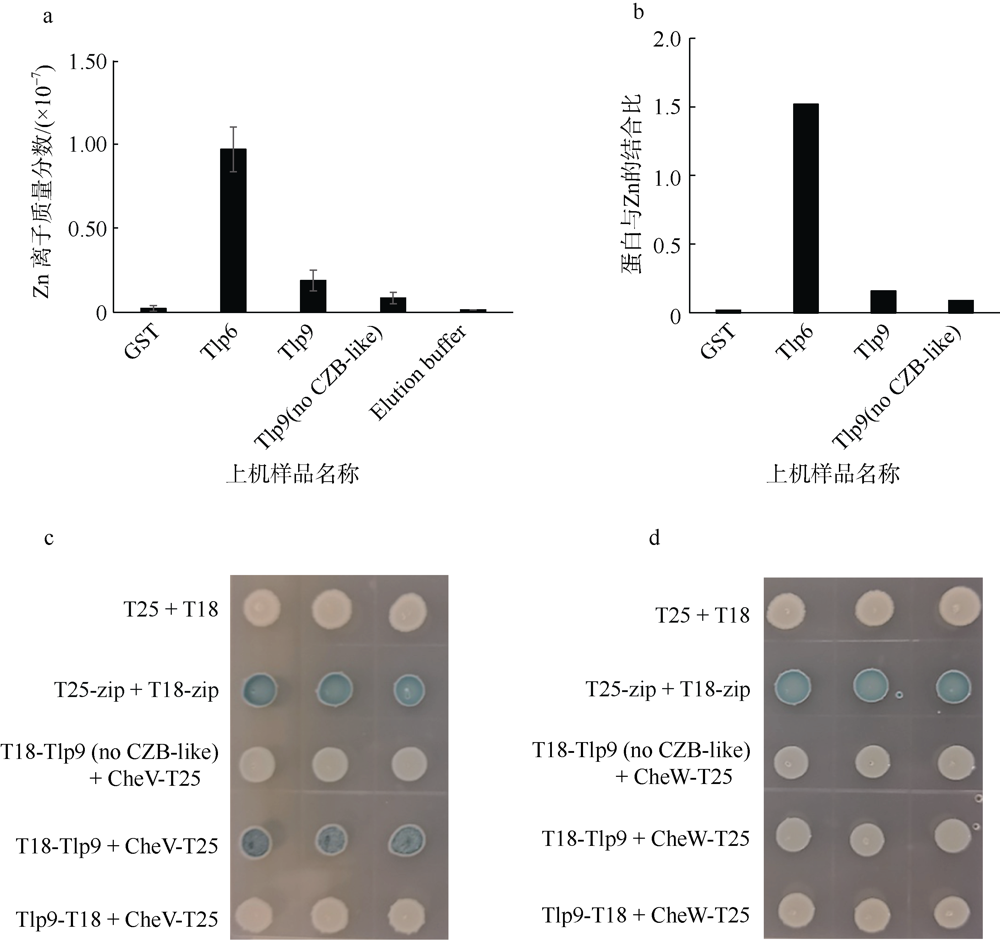
Fig. 5
Study on Zn-binding capacity and potential function of CZB-like domain (a) Zn concentration measure by ICP-mass; (b) Using the average Zn concentration and average protein concentration, the binding ratio of protein and Zn is calculated; (c and d) To investigate whether the CZB-like domain of Tlp9 can affect the interaction with CheV or CheW by bacterial two-hybrid experiments. The shade of color in Figure C and D represents the strength of protein interaction"
| [1] | 王风平, 周悦恒, 张新旭, 等, 2013. 深海微生物多样性[J]. 生物多样性, 21(4):445-455. |
| WANG FENGPING, ZHOU YUEHENG, ZHANG XINXU, et al, 2013. Biodiversity of deep-sea microorganisms[J]. Biodiversity Science, 21(4):445-455 (in Chinese with English abstract). | |
| [2] | 张云怡, 曾令兵, 郭晓奎, 等, 2011. 细菌趋化过程中信号转导系统研究[J]. 中国微生态学杂志, 23(1):93-96 (in Chinese). |
| [3] | 臧扬, 高贝乐, 2017. 深海热液口Epsilon-变形菌的物种多样性与环境适应机理[J]. 微生物学报, 57(9):1392-1399. |
| ZANG YANG, GAO BEILE, 2017. Biodiversity and environmental adaptation of deep-sea hydrothermal vent Epsilon- proteobacteria[J]. Acta Microbiologica Sinica, 57(9):1392-1399 (in Chinese with English abstract). | |
| [4] |
ALEXANDER R P, LOWENTHAL A C, HARSHEY R M, et al, 2010. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network[J]. Trends in Microbiology, 18(11):494-503.
doi: 10.1016/j.tim.2010.07.004 pmid: 20832320 |
| [5] |
ALTSCHUL S F, MADDEN T L, SCHÄFFER A A, et al, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs[J]. Nucleic Acids Research, 25(17):3389-3402.
pmid: 9254694 |
| [6] |
BATTESTI A, BOUVERET E, 2012. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli[J]. Methods, 58(4):325-334.
pmid: 22841567 |
| [7] |
CAMPBELL B J, ENGEL A S, PORTER M L, et al, 2006. The versatile ε-proteobacteria: key players in sulphidic habitats[J]. Nature Reviews Microbiology, 4(6):458-468.
pmid: 16652138 |
| [8] |
CASSIDY C K, HIMES B A, ALVAREZ F J, et al, 2015. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling[J]. eLife, 4: e08419.
doi: 10.7554/eLife.08419 pmid: 26583751 |
| [9] |
COLLINS K D, ANDERMANN T M, DRAPER J, et al, 2016. The Helicobacter pylori CZB cytoplasmic chemoreceptor TlpD forms an autonomous polar chemotaxis signaling complex that mediates a tactic response to oxidative stress[J]. Journal of Bacteriology, 198(11):1563-1575.
doi: 10.1128/JB.00071-16 pmid: 27002127 |
| [10] |
CROOKS G E, HON G, CHANDONIA J M, et al, 2004. WebLogo: a sequence logo generator[J]. Genome Research, 14(6):1188-1190.
pmid: 15173120 |
| [11] |
DELALEZ N J, WADHAMS G H, ROSSER G, et al, 2010. Signal-dependent turnover of the bacterial flagellar switch protein FliM[J]. Proceedings of the National Academy of Sciences of the United States of America, 107(25):11347-11351.
pmid: 20498085 |
| [12] |
ELLIOTT K T, ZHULIN I B, STUCKEY J A, et al, 2009. Conserved residues in the HAMP domain define a new family of proposed bipartite energy taxis receptors[J]. Journal of Bacteriology, 191(1):375-387.
pmid: 18952801 |
| [13] |
GIBSON D G, YOUNG L, CHUANG R Y, et al, 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases[J]. Nature Methods, 6(5):343-345.
doi: 10.1038/nmeth.1318 pmid: 19363495 |
| [14] |
GIOVANNELLI D, FERRIERA S, JOHNSON J, et al, 2011. Draft genome sequence of Caminibacter mediatlanticus strain TB-2 T, an epsilonproteobacterium isolated from a deep-sea hydrothermal vent [J]. Standards in Genomic Sciences, 5(1):135-143.
doi: 10.4056/sigs.2094859 pmid: 22180817 |
| [15] |
GROSCHE A, SEKARAN H, PÉREZ-RODRÍGUEZ I, et al, 2015. Cetia pacifica gen. nov., sp. nov., a chemolithoautotrophic, thermophilic, nitrate-ammonifying bacterium from a deep-sea hydrothermal vent[J]. International Journal of Systematic and Evolutionary Microbiology, 65: 1144-1150.
pmid: 25604337 |
| [16] |
GUMEROV V M, ORTEGA D R, ADEBALI O, et al, 2020. MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems[J]. Nucleic Acids Research, 48(D1):D459-D464.
pmid: 31754718 |
| [17] |
INAGAKI F, TAKAI K, KOBAYASHI H, et al, 2003. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur- oxidizing ε-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa trough[J]. International Journal of Systematic and Evolutionary Microbiology, 53(Pt 6):1801-1805.
pmid: 14657107 |
| [18] | KARATAN E, SAULMON M M, BUNN M W, et al, 2001. Phosphorylation of the response regulator CheV is required for adaptation to attractants during Bacillus subtilis chemotaxis[J]. Journal of Biological Chemistry, 276(47):43618-43626. |
| [19] |
KUMAR S, STECHER G, TAMURA K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 33(7):1870-1874.
pmid: 27004904 |
| [20] |
LARKIN M A, BLACKSHIELDS G, BROWN N P, et al, 2007. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 23(21):2947-2948.
pmid: 17846036 |
| [21] |
LERTSETHTAKARN P, OTTEMANN K M, HENDRIXSON D R, 2011. Motility and chemotaxis in Campylobacter and Helicobacter[J]. Annual Review of Microbiology, 65: 389-410.
pmid: 21939377 |
| [22] |
LETUNIC I, BORK P, 2018. 20 years of the SMART protein domain annotation resource[J]. Nucleic Acids Research, 46(D1):D493-D496.
doi: 10.1093/nar/gkx922 pmid: 29040681 |
| [23] |
LIU JUN, HU BO, MORADO D R, et al, 2012. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells[J]. Proceedings of the National Academy of Sciences of the United States of America, 109(23):E1481-E1488.
pmid: 22556268 |
| [24] |
MCNICHOL J, STRYHANYUK H, SYLVA S P, et al, 2018. Primary productivity below the seafloor at deep-sea hot springs[J]. Proceedings of the National Academy of Sciences of the United States of America, 115(26):6756-6761.
pmid: 29891698 |
| [25] |
MEYER J L, HUBER J A, 2014. Strain-level genomic variation in natural populations of Lebetimonas from an erupting deep-sea volcano[J]. The ISME Journal, 8(4):867-880.
pmid: 24257443 |
| [26] | NA S I, KIM Y O, YOON S H, et al, 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction[J]. Journal of Microbiology, 56(4):280-285. |
| [27] |
NAKAGAWA S, TAKAI K, INAGAKI F, et al, 2005. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ε-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa trough[J]. International Journal of Systematic and Evolutionary Microbiology, 55(Pt 2):925-933.
pmid: 15774687 |
| [28] |
ORTEGA D R, ZHULIN I B, 2016. Evolutionary genomics suggests that CheV is an additional adaptor for accommodating specific chemoreceptors within the chemotaxis signaling complex[J]. PLoS Computational Biology, 12(2):e1004723.
doi: 10.1371/journal.pcbi.1004723 pmid: 26844549 |
| [29] |
PARKINSON J S, HAZELBAUER G L, FALKE J J, 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update[J]. Trends in Microbiology, 23(5):257-266.
doi: 10.1016/j.tim.2015.03.003 pmid: 25834953 |
| [30] |
PIÑAS G E, FRANK V, VAKNIN A, et al, 2016. The source of high signal cooperativity in bacterial chemosensory arrays[J]. Proceedings of the National Academy of Sciences of the United States of America, 113(12):3335-3340.
doi: 10.1073/pnas.1600216113 pmid: 26951681 |
| [31] |
PITTMAN M S, GOODWIN M, KELLY D J, 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation[J]. Microbiology, 147(Pt 9):2493-2504.
doi: 10.1099/00221287-147-9-2493 pmid: 11535789 |
| [32] |
PORTER S L, WADHAMS G H, ARMITAGE J P, 2011. Signal processing in complex chemotaxis pathways[J]. Nature Reviews Microbiology, 9(3):153-165.
doi: 10.1038/nrmicro2505 pmid: 21283116 |
| [33] |
RAO C V, GLEKAS G D, ORDAL G W, 2008. The three adaptation systems of Bacillus subtilis chemotaxis[J]. Trends in Microbiology, 16(10):480-487.
doi: 10.1016/j.tim.2008.07.003 pmid: 18774298 |
| [34] |
ROSARIO M M, FREDRICK K L, ORDAL G W, et al, 1994. Chemotaxis in Bacillus subtilis requires either of two functionally redundant CheW homologs[J]. Journal of Bacteriology, 176(9):2736-2739.
doi: 10.1128/jb.176.9.2736-2739.1994 pmid: 8169224 |
| [35] |
SCHÄFFER A A, ARAVIND L, MADDEN T L, et al, 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements[J]. Nucleic Acids Research, 29(14):2994-3005.
pmid: 11452024 |
| [36] |
SMITH J L, CAMPBELL B J, HANSON T E, et al, 2008. Nautilia profundicola sp. nov., a thermophilic, sulfur-reducing epsilonproteobacterium from deep-sea hydrothermal vents[J]. International Journal of Systematic and Evolutionary Microbiology, 58(Pt 7):1598-1602.
doi: 10.1099/ijs.0.65435-0 pmid: 18599701 |
| [37] |
SNELLING W J, MATSUDA M, MOORE J E, et al, 2005. Campylobacter jejuni[J]. Letters in Applied Microbiology, 41(4):297-302.
doi: 10.1111/j.1472-765X.2005.01788.x pmid: 16162134 |
| [38] |
STOCKER R, SEYMOUR J R, 2012. Ecology and physics of bacterial chemotaxis in the ocean[J]. Microbiology and Molecular Biology Reviews, 76(4):792-812.
doi: 10.1128/MMBR.00029-12 pmid: 23204367 |
| [39] |
TAKAI K, NEALSON K H, HORIKOSHI K, 2004. Hydrogenimonas thermophila gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing chemolithoautotroph within the ε-Proteobacteria, isolated from a black smoker in a central Indian ridge hydrothermal field[J]. International Journal of Systematic and Evolutionary Microbiology, 54(Pt 1):25-32.
doi: 10.1099/ijs.0.02787-0 pmid: 14742455 |
| [40] |
WADHAMS G H, ARMITAGE J P, 2004. Making sense of it all: bacterial chemotaxis[J]. Nature Reviews Molecular Cell Biology, 5(12):1024-1037.
pmid: 15573139 |
| [41] |
WAITE D W, VANWONTERGHEM I, RINKE C, et al, 2017. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.)[J]. Frontiers in Microbiology, 8: 682.
doi: 10.3389/fmicb.2017.00682 pmid: 28484436 |
| [42] |
WUICHET K, ZHULIN I B, 2010. Origins and diversification of a complex signal transduction system in prokaryotes[J]. Science Signaling, 3(128): ra50.
doi: 10.1126/scisignal.3128eg5 pmid: 20587802 |
| [43] |
ZAHRINGER F, LACANNA E, JENAL U, et al, 2013. Structure and signaling mechanism of a zinc-sensory diguanylate cyclase[J]. Structure, 21(7):1149-1157.
pmid: 23769666 |
| [44] |
ZAUTNER A E, TAREEN A M, GROß U, et al, 2012. Chemotaxis in Campylobacter jejuni[J]. European Journal of Microbiology and Immunology, 2(1):24-31.
doi: 10.1556/EuJMI.2.2012.1.5 pmid: 24611118 |
| [45] |
ZIMMERMANN L, STEPHENS A, NAM S Z, et al, 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core[J]. Journal of Molecular Biology, 430(15):2237-2243.
doi: 10.1016/j.jmb.2017.12.007 pmid: 29258817 |
| [1] | WANG Junfeng, ZHOU Wenying, TIAN Xinpeng, LIU Yonghong. Antiviral chromones from the deep-sea hydrothermal-vent-derived Fusarium sp. SCSIO 06196 [J]. Journal of Tropical Oceanography, 2022, 41(5): 189-193. |
| [2] | CONG Mengjing, HU Yiwei, ZHAO Kai, ZHANG Xiaoyong, LIU Yonghong, WANG Junfeng. Study on the polyketides from a cold-seep-derived Talaromyces helicus SCSIO41311 [J]. Journal of Tropical Oceanography, 2022, 41(5): 117-120. |
| [3] | YE Ziqing, ZHAO Xiangdan, CAI Bingna, WAN Peng, CHEN Hua, PAN Jianyu. Oyster hydrolysates alleviate 5-fluorouracil-induced intestinal mucosal injury on S180 tumor-bearing mice* [J]. Journal of Tropical Oceanography, 2022, 41(5): 105-116. |
| [4] | PAN Xiaolan, LIU Huiru, XU Meng, XU Hanzhi, ZHANG Hua, HE Maoxian. Preliminary study on immunity function of aquaporin AQP4 in the pearl oyster Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2021, 40(2): 83-89. |
| [5] | WANG Xiaomin, DONG Lu, ZHANG Jifu, ZHANG Yun, HU Yunfeng. The effects of different cell breaking methods on the asymmetrical hydrolysis of phenylethyl acetate using intracellular proteases of Bacillus sp. DL-2 [J]. Journal of Tropical Oceanography, 2021, 40(2): 39-48. |
| [6] | Yanhong TAN, Jixing LI, Xiuping LIN, Bin YANG, Yonghong LIU, Yunqiu LI. Study on the secondary metabolites from the South China Sea soft coral-derived fungus Eupenicillium sp. DX-SER3 (KC871024) [J]. Journal of Tropical Oceanography, 2019, 38(2): 43-47. |
| [7] | Xuanyu ZHAO, Fei SHI, Xiaoxue WANG. Characterization of a ParE-ParD toxin-antitoxin TA system (SO_A0087-A0088) on a megaplasmid of Shewanella oneidensis [J]. Journal of Tropical Oceanography, 2018, 37(6): 104-111. |
| [8] | WANG Zhao-kai, YI Zhi-wei, LIU Yang, CHAN Zhu-hua, ZENG Run-ying. Isolation, identification and characterization of an esterase-producing marine moderately halophilic strain Halomonas sp.LYG1-1 [J]. Journal of Tropical Oceanography, 2014, 33(1): 69-73. |
| [9] | ZHANG Yun, HUANG Kai-xuan, OU Lin-jian, QIN Xian-ling, WANG Zhao-hui, HUANG Dao-jian, QI Yu-zao. Distributions of urea concentration and urease activity in the Daya Bay [J]. Journal of Tropical Oceanography, 2014, 33(1): 90-96. |
|
||