Journal of Tropical Oceanography ›› 2023, Vol. 42 ›› Issue (2): 78-86.doi: 10.11978/2022108CSTR: 32234.14.2022108
Previous Articles Next Articles
Discovery and verification of SNP in Acanthopagrus latus
ZHENG Guobin( ), ZHAO Hongbo, HUANG Liangmin, ZHANG Jing, LIU Xiande(
), ZHAO Hongbo, HUANG Liangmin, ZHANG Jing, LIU Xiande( )
)
- Fishery College of Jimei University, Key Laboratory of Mariculture for the East China Sea, Ministry of Agriculture and Rural Affairs, Xiamen 361021, China
-
Received:2022-05-13Revised:2022-07-09Online:2023-03-10Published:2022-07-14 -
Contact:LIU Xiande. email:xdliu@jmu.edu -
Supported by:National Key R&D Program of China(2018YFD0901404); Evaluation of the Release Effect of Acanthopagrus latus in Xiamen Bay(S20166)
Cite this article
ZHENG Guobin, ZHAO Hongbo, HUANG Liangmin, ZHANG Jing, LIU Xiande. Discovery and verification of SNP in Acanthopagrus latus[J].Journal of Tropical Oceanography, 2023, 42(2): 78-86.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 2
The results of re-sequence data quality"
| 样品 | 原始数据/Gb | 过滤数据/Gb | 比对率/% | 测序深度/× | Q20/% | Q30/% | GC含量/% | 样品 | 原始数据/Gb | 过滤数据/Gb | 比对率/% | 测序深度/× | Q20/% | Q30/% | GC含量/% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Y01 | 4.71 | 4.71 | 95.90 | 6.15 | 97.34 | 92.78 | 42.69 | Y26 | 4.79 | 4.79 | 97.00 | 6.33 | 97.33 | 92.77 | 42.80 |
| Y02 | 5.00 | 4.99 | 95.49 | 6.52 | 97.39 | 92.88 | 42.69 | Y27 | 4.59 | 4.59 | 96.96 | 6.09 | 97.27 | 92.52 | 42.93 |
| Y03 | 4.76 | 4.76 | 96.20 | 6.22 | 97.43 | 92.94 | 42.61 | Y28 | 4.63 | 4.63 | 96.76 | 6.10 | 96.56 | 91.10 | 42.78 |
| Y04 | 4.55 | 4.55 | 96.83 | 6.03 | 97.49 | 93.13 | 42.73 | Y29 | 4.74 | 4.74 | 96.74 | 6.27 | 97.34 | 92.84 | 42.83 |
| Y05 | 4.58 | 4.58 | 96.38 | 6.06 | 97.43 | 93.03 | 42.76 | Y30 | 4.48 | 4.48 | 96.96 | 5.96 | 97.54 | 93.17 | 42.66 |
| Y06 | 4.46 | 4.46 | 96.46 | 5.92 | 97.44 | 93.02 | 42.69 | Y31 | 4.85 | 4.85 | 97.2 | 6.41 | 97.47 | 93.06 | 42.66 |
| Y07 | 4.71 | 4.71 | 97.03 | 6.25 | 97.45 | 93.06 | 42.76 | Y32 | 4.17 | 4.17 | 97.10 | 5.59 | 97.26 | 92.60 | 42.75 |
| Y08 | 4.79 | 4.79 | 96.69 | 6.32 | 97.21 | 92.52 | 42.80 | Y33 | 4.98 | 4.98 | 97.27 | 6.55 | 97.27 | 92.68 | 42.99 |
| Y09 | 4.86 | 4.86 | 97.36 | 6.43 | 97.34 | 92.82 | 43.18 | Y34 | 3.69 | 3.69 | 96.8 | 5.00 | 97.29 | 92.64 | 42.96 |
| Y10 | 4.78 | 4.77 | 96.60 | 6.30 | 97.19 | 92.46 | 42.87 | Y35 | 4.92 | 4.92 | 97.33 | 6.48 | 97.12 | 92.31 | 43.03 |
| Y11 | 4.43 | 4.43 | 96.56 | 5.88 | 96.97 | 92.00 | 42.83 | Y36 | 4.86 | 4.86 | 97.64 | 6.41 | 97.23 | 92.58 | 42.95 |
| Y12 | 4.59 | 4.59 | 96.60 | 6.06 | 97.09 | 92.23 | 42.95 | Y37 | 4.88 | 4.88 | 97.32 | 6.45 | 97.04 | 92.13 | 42.79 |
| Y13 | 4.63 | 4.63 | 96.87 | 6.13 | 97.26 | 92.64 | 42.78 | Y38 | 4.84 | 4.84 | 96.84 | 6.35 | 97.25 | 92.59 | 42.74 |
| Y14 | 4.82 | 4.82 | 96.55 | 6.32 | 97.08 | 92.24 | 42.71 | Y39 | 4.59 | 4.59 | 96.43 | 6.07 | 97.28 | 92.65 | 42.85 |
| Y15 | 4.63 | 4.63 | 96.57 | 6.12 | 97.22 | 92.49 | 42.77 | Y40 | 4.23 | 4.23 | 97.54 | 5.66 | 97.36 | 92.82 | 42.95 |
| Y16 | 4.70 | 4.69 | 96.80 | 6.20 | 97.24 | 92.61 | 42.97 | Y41 | 4.73 | 4.73 | 97.65 | 6.26 | 97.49 | 93.13 | 43.01 |
| Y17 | 4.93 | 4.93 | 96.95 | 6.50 | 97.17 | 92.43 | 42.91 | Y42 | 4.37 | 4.37 | 96.58 | 5.77 | 97.26 | 92.61 | 43.08 |
| Y18 | 4.33 | 4.33 | 97.05 | 5.79 | 97.10 | 92.22 | 42.81 | Y43 | 4.90 | 4.90 | 97.08 | 6.45 | 97.42 | 92.99 | 43.01 |
| Y19 | 4.85 | 4.85 | 96.91 | 6.40 | 97.25 | 92.59 | 42.95 | Y44 | 4.82 | 4.81 | 97.36 | 6.36 | 97.49 | 93.09 | 42.99 |
| Y20 | 4.52 | 4.52 | 96.45 | 5.96 | 97.33 | 92.76 | 42.75 | Y45 | 4.95 | 4.95 | 97.14 | 6.52 | 97.50 | 93.16 | 43.02 |
| Y21 | 4.79 | 4.79 | 97.00 | 6.32 | 97.20 | 92.46 | 42.73 | Y46 | 4.61 | 4.61 | 97.09 | 5.85 | 97.46 | 93.06 | 42.68 |
| Y22 | 4.84 | 4.84 | 97.00 | 6.36 | 97.54 | 93.21 | 42.54 | Y47 | 4.75 | 4.75 | 96.72 | 6.27 | 97.42 | 92.98 | 42.86 |
| Y23 | 4.71 | 4.71 | 96.41 | 6.17 | 96.71 | 91.43 | 42.72 | Y48 | 4.50 | 4.50 | 96.85 | 5.96 | 97.44 | 93.02 | 42.90 |
| Y24 | 4.58 | 4.58 | 96.84 | 6.08 | 97.25 | 92.56 | 42.75 | Y49 | 4.42 | 4.42 | 97.43 | 5.90 | 97.17 | 92.46 | 43.33 |
| Y25 | 4.75 | 4.75 | 97.22 | 6.27 | 97.33 | 92.74 | 42.80 | Y50 | 4.88 | 4.88 | 97.36 | 6.44 | 97.38 | 92.88 | 43.03 |
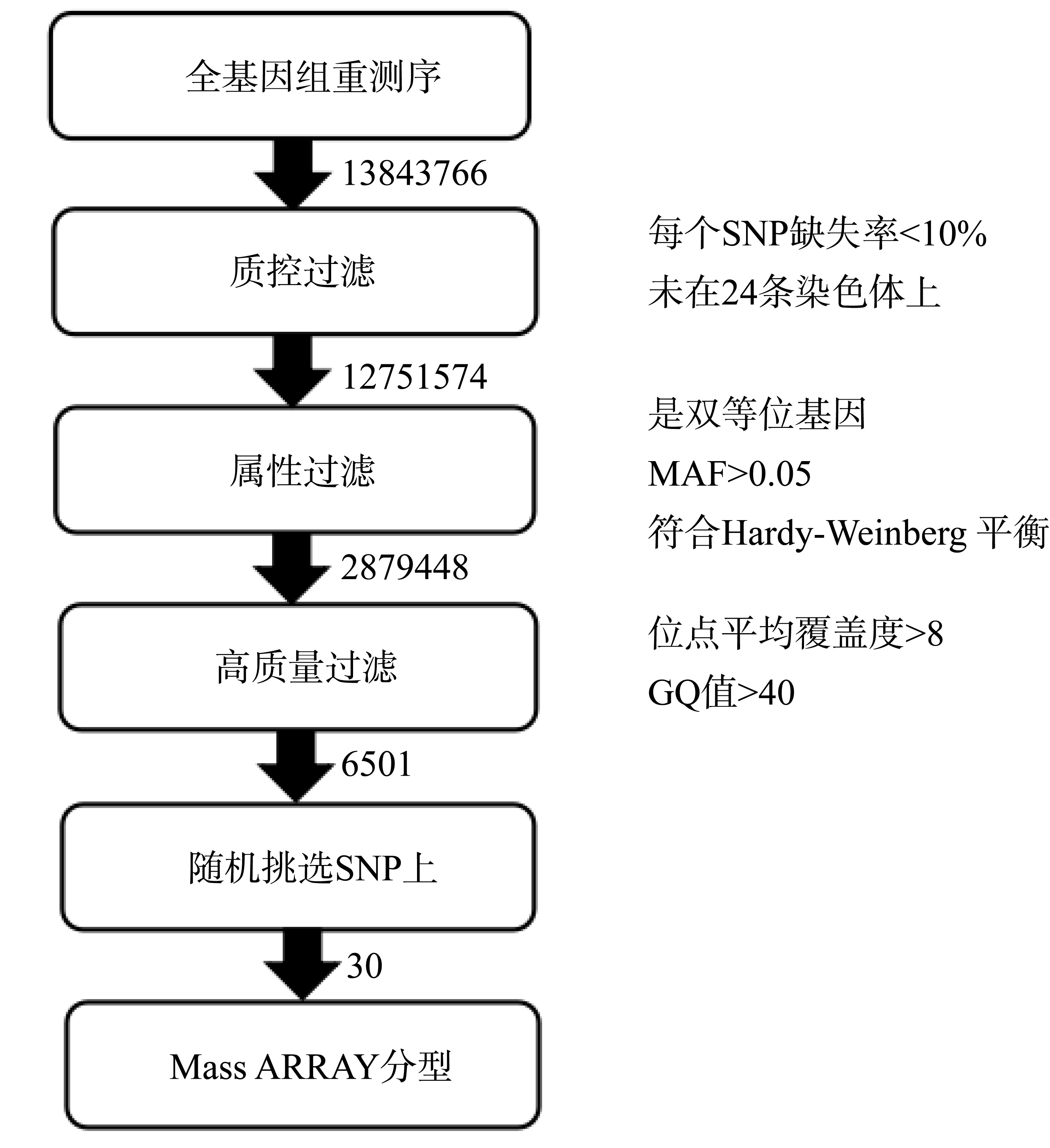
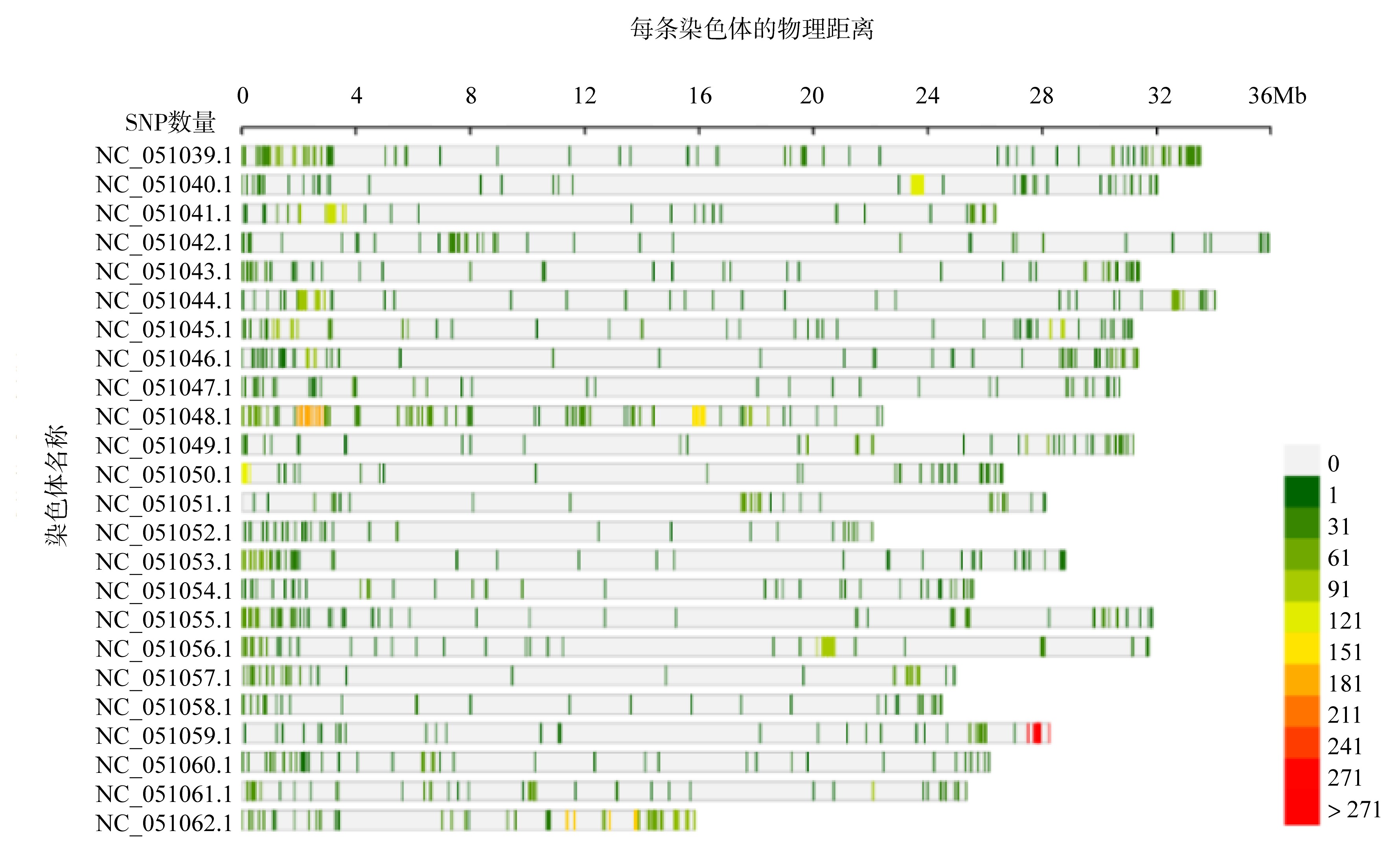
Tab. 3
The results of SNP discovery and filtering in re-sequenced Acanthopagrus latus genome"
| 染色体名称 | SNP初始数 | 质控过滤后SNP数 | 属性过滤后SNP数 | 高质量过滤后SNP数 |
|---|---|---|---|---|
| NC_051039.1 | 692976 | 632996 | 144274 | 364 |
| NC_051040.1 | 646881 | 595158 | 134036 | 221 |
| NC_051041.1 | 566757 | 522152 | 117157 | 325 |
| NC_051042.1 | 690338 | 640940 | 142536 | 172 |
| NC_051043.1 | 606086 | 565937 | 129160 | 157 |
| NC_051044.1 | 697405 | 638444 | 144000 | 265 |
| NC_051045.1 | 589449 | 548080 | 124463 | 366 |
| NC_051046.1 | 615425 | 574376 | 130360 | 228 |
| NC_051047.1 | 622596 | 577032 | 131271 | 203 |
| NC_051048.1 | 476295 | 405575 | 97778 | 820 |
| NC_051049.1 | 629215 | 586030 | 132799 | 263 |
| NC_051050.1 | 574454 | 532521 | 120384 | 237 |
| NC_051051.1 | 567923 | 530390 | 119320 | 139 |
| NC_051052.1 | 462308 | 429823 | 94073 | 109 |
| NC_051053.1 | 538049 | 501748 | 110871 | 172 |
| NC_051054.1 | 535520 | 495987 | 112649 | 173 |
| NC_051055.1 | 613575 | 567469 | 128275 | 221 |
| NC_051056.1 | 669814 | 618189 | 137376 | 256 |
| NC_051057.1 | 520747 | 477109 | 104765 | 187 |
| NC_051058.1 | 509767 | 475642 | 106603 | 97 |
| NC_051059.1 | 583650 | 535046 | 121219 | 370 |
| NC_051060.1 | 563226 | 523705 | 117774 | 169 |
| NC_051061.1 | 518795 | 479194 | 110565 | 299 |
| NC_051062.1 | 329951 | 298031 | 67740 | 688 |
| 未知序列 | 22564 | 0 | 0 | 0 |
| 合计 | 13843766 | 12751574 | 2879448 | 6501 |
Tab. 5
Genotyping comparison between genome re-sequencing and MassARRAY"
| SNP | 染色体名称 | 位置 | MassARRAY分型检测 | 全基因组重测序 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 检测结果 | 检出率/% | MF | MAF | 检测结果 | 检出率/% | MF | MAF | |||
| Shq_178_1 | NC_051039.1 | 33527645 | CC、CG | 85 | 0.85 | 0.15 | CC、CG | 95 | 0.74 | 0.26 |
| Shq_178_2 | NC_051040.1 | 32016498 | TT | 100 | 1.00 | 0 | TT、TG | 100 | 0.88 | 0.13 |
| Shq_178_3 | NC_051041.1 | 20806265 | TT、TG | 100 | 0.95 | 0.05 | TT、TG | 100 | 0.75 | 0.25 |
| Shq_178_6 | NC_051042.1 | 30329 | AA、AG | 90 | 0.69 | 0.31 | AA、AG | 95 | 0.63 | 0.37 |
| Shq_178_8 | NC_051042.1 | 7311706 | TT、TC | 100 | 0.95 | 0.05 | TT、TC | 100 | 0.95 | 0.05 |
| Shq_178_19 | NC_051048.1 | 22404460 | AA | 95 | 1.00 | 0 | AA、AG | 90 | 0.92 | 0.08 |
| Shq_178_26 | NC_051050.1 | 34885 | CC | 95 | 1.00 | 0 | CC、CA | 95 | 0.92 | 0.08 |
| Shq_178_39 | NC_051050.1 | 38757 | GG | 100 | 1.00 | 0 | GG、GA | 90 | 0.94 | 0.06 |
| Shq_178_40 | NC_051050.1 | 42037 | TT | 100 | 1.00 | 0 | TT、TC | 95 | 0.87 | 0.13 |
| Shq_178_48 | NC_051050.1 | 59154 | TT、TC | 85 | 0.91 | 0.09 | TT、TC | 100 | 0.65 | 0.35 |
| Shq_178_60 | NC_051050.1 | 74750 | GG | 100 | 1.00 | 0 | GG、GA | 90 | 0.94 | 0.06 |
| Shq_178_63 | NC_051050.1 | 75488 | GG | 100 | 1.00 | 0 | GG、GC | 95 | 0.95 | 0.05 |
| Shq_178_65 | NC_051050.1 | 76081 | AA | 100 | 1.00 | 0 | AA、AC | 85 | 0.88 | 0.12 |
| Shq_178_70 | NC_051050.1 | 87063 | GG | 100 | 1.00 | 0 | GG、GA | 90 | 0.89 | 0.11 |
| Shq_178_72 | NC_051050.1 | 93481 | CC、CT | 100 | 0.98 | 0.03 | CC、CT | 100 | 0.95 | 0.05 |
| Shq_178_73 | NC_051050.1 | 96282 | CC | 100 | 1.00 | 0 | CC、CA | 95 | 0.82 | 0.18 |
| Shq_178_77 | NC_051050.1 | 101791 | CC | 100 | 1.00 | 0 | CC、CT | 80 | 0.94 | 0.06 |
| Shq_178_86 | NC_051050.1 | 106638 | GG、GA | 100 | 0.97 | 0.03 | GG、GA | 80 | 0.97 | 0.03 |
| Shq_178_92 | NC_051050.1 | 128900 | AA | 100 | 1.00 | 0 | AA、AG | 100 | 0.93 | 0.08 |
| Shq_178_96 | NC_051050.1 | 142395 | GG | 100 | 1.00 | 0 | GG、GA | 90 | 0.92 | 0.08 |
| Shq_178_104 | NC_051052.1 | 1588997 | AA | 100 | 1.00 | 0 | AA、AC | 100 | 0.75 | 0.25 |
| Shq_178_112 | NC_051053.1 | 1538 | GG | 100 | 1.00 | 0 | AA、AG、GG | 95 | 0.53 | 0.47 |
| Shq_178_138 | NC_051053.1 | 8936396 | AA | 100 | 1.00 | 0 | AA、AG | 95 | 0.76 | 0.24 |
| Shq_178_149 | NC_051056.1 | 31720087 | CC、CT | 100 | 0.95 | 0.05 | CC、CT | 90 | 0.86 | 0.14 |
| Shq_178_150 | NC_051056.1 | 31723482 | TT | 100 | 1.00 | 0 | TT、TC | 95 | 0.89 | 0.11 |
| Shq_178_157 | NC_051058.1 | 24478462 | AA、AG | 100 | 0.95 | 0.05 | AA、AC | 100 | 0.95 | 0.05 |
| Shq_178_158 | NC_051058.1 | 24478507 | CC、CT | 95 | 0.97 | 0.03 | CC、CT | 100 | 0.95 | 0.05 |
| Shq_178_171 | NC_051060.1 | 26149536 | GG | 100 | 1.00 | 0 | GG、GC | 100 | 0.58 | 0.43 |
| Shq_178_172 | NC_051061.1 | 336649 | TT | 100 | 1.00 | 0 | TT、TG | 100 | 0.55 | 0.45 |
| Shq_178_173 | NC_051061.1 | 22105514 | GG | 100 | 1.00 | 0 | GG、GA | 100 | 0.55 | 0.45 |
| [1] |
江兴龙, 黄永春, 黄良敏, 等, 2013. 厦门湾黄鳍鲷增殖放流效果的评估[J]. 集美大学学报(自然科学版), 18(3): 161-166.
|
|
|
|
| [2] |
吴利娜, 张凝鋆, 孙松, 等, 2021. 微卫星分子标记技术在大黄鱼增殖放流效果评估中的应用[J]. 中国水产科学, 28(9): 1100-1108.
|
|
|
|
| [3] |
杨习文, 刘熠, 薛向平, 等, 2020. 基于微卫星标记的长江江苏段鲢(Hypophthalmichthys molitrix)增殖放流资源贡献率的评估[J]. 湖泊科学, 32(4): 1154-1164.
|
|
|
|
| [4] |
赵雨, 2021. 基于微卫星标记的日本对虾增殖放流效果评价及群体遗传学研究[D]. 天津农学院.
|
|
|
|
| [5] |
朱克诚, 宋岭, 刘宝锁, 等, 2020. 黄鳍棘鲷家系亲缘关系鉴定[J]. 水产学报, 44(3): 351-357.
|
|
|
|
| [6] |
doi: 10.1111/eva.12711 |
| [7] |
|
| [8] |
doi: 10.1093/bioinformatics/btx692 |
| [9] |
doi: 10.3389/fgene.2021.671650 |
| [10] |
|
| [11] |
|
| [12] |
doi: 10.1016/j.aquaculture.2018.06.019 |
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
doi: 10.1093/bioinformatics/btp352 pmid: 19505943 |
| [17] |
doi: 10.1016/j.aquaculture.2015.11.001 |
| [18] |
doi: 10.1101/gr.107524.110 pmid: 20644199 |
| [19] |
doi: 10.1007/s00414-016-1490-5 |
| [20] |
doi: 10.1371/journal.pone.0070051 |
| [21] |
|
| [22] |
doi: 10.1007/s100380170047 pmid: 11501945 |
| [23] |
|
| [24] |
doi: 10.1007/s00414-013-0879-7 pmid: 23736940 |
| [25] |
doi: 10.1371/journal.pone.0058700 |
| [26] |
doi: 10.3390/ijms14035694 pmid: 23481633 |
| [27] |
doi: 10.2983/0730-8000(2008)27[489:ILPIOA]2.0.CO;2 |
| [28] |
doi: 10.1111/1755-0998.12503 pmid: 26849107 |
| [29] |
doi: 10.1007/s10126-019-09935-5 pmid: 31938972 |
| [30] |
doi: 10.1016/j.aquaculture.2017.11.014 |
| [1] | HUANG Jing, PAN Xiaolan, XU Meng, LIU Wenguang, ZHANG Hua, HE Maoxian. Morphological and SNP markers for analysis of genetic structure of hybrid progeny and their parental populations of Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2019, 38(6): 80-89. |
| [2] | Jialin AI, Zhimin LI, Jianyong LIU, Yuchun SHEN. Single nucleotide polymorphisms of myostation gene and its association with growth traits in Haliotis diversicolor supertexta [J]. Journal of Tropical Oceanography, 2019, 38(2): 78-85. |
|
||