Journal of Tropical Oceanography ›› 2023, Vol. 42 ›› Issue (5): 92-103.doi: 10.11978/2022250CSTR: 32234.14.2022250
• Marine Biology • Previous Articles Next Articles
Transcriptomic analysis of fatty acid metabolism in the Thalassiosira pseudonana under low salinity stress
SUN Wenjie1,2( ), LI Jiamin1,2, WANG Hualong3, MI Tiezhu1,2,4(
), LI Jiamin1,2, WANG Hualong3, MI Tiezhu1,2,4( ), ZHEN Yu1,2,4
), ZHEN Yu1,2,4
- 1. College of Environmental Science and Engineering, Ocean University of China, Qingdao 266100, China
2. Key Laboratory of Marine Environment and Ecology Ministry of Education, Ocean University of China, Qingdao 266100, China
3. College of Marine Life Science, Ocean University of China, Qingdao 266003, China
4. Laboratory for Marine Ecology and Environmental Science Qingdao National Laboratory for Marine Science and Technology, Qingdao 266071, China
-
Received:2022-12-01Revised:2023-01-12Online:2023-09-10Published:2023-02-22 -
Supported by:Qingdao National Laboratory of Marine Science and Technology(2021QNLM040001); National Natural Science Foundation of China(41976133)
Cite this article
SUN Wenjie, LI Jiamin, WANG Hualong, MI Tiezhu, ZHEN Yu. Transcriptomic analysis of fatty acid metabolism in the Thalassiosira pseudonana under low salinity stress[J].Journal of Tropical Oceanography, 2023, 42(5): 92-103.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
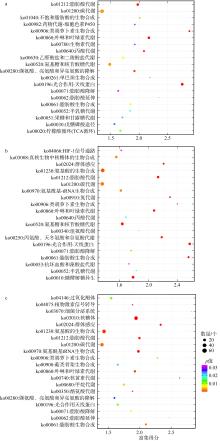
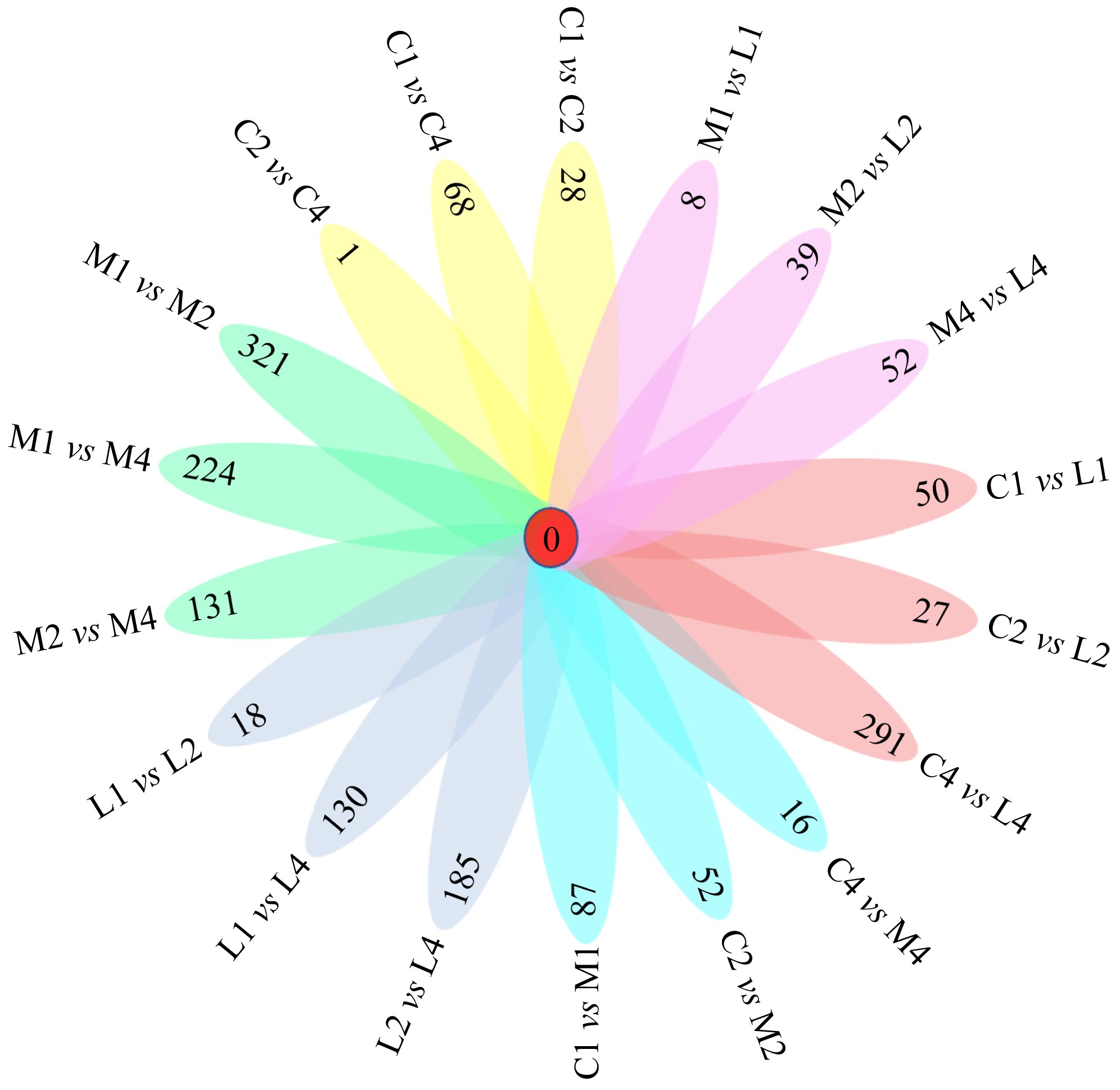
Fig. 1
Shared expressed genes between two groups. C1/C2/C4: the control group on day one, day two and day four; M1/M2/M4: the middle salinity group on day one, day two and day four; L1/L2/L4: the low salinity group on day one, day two and day four. Number in each oval represents the count of shared expressed genes between two groups"
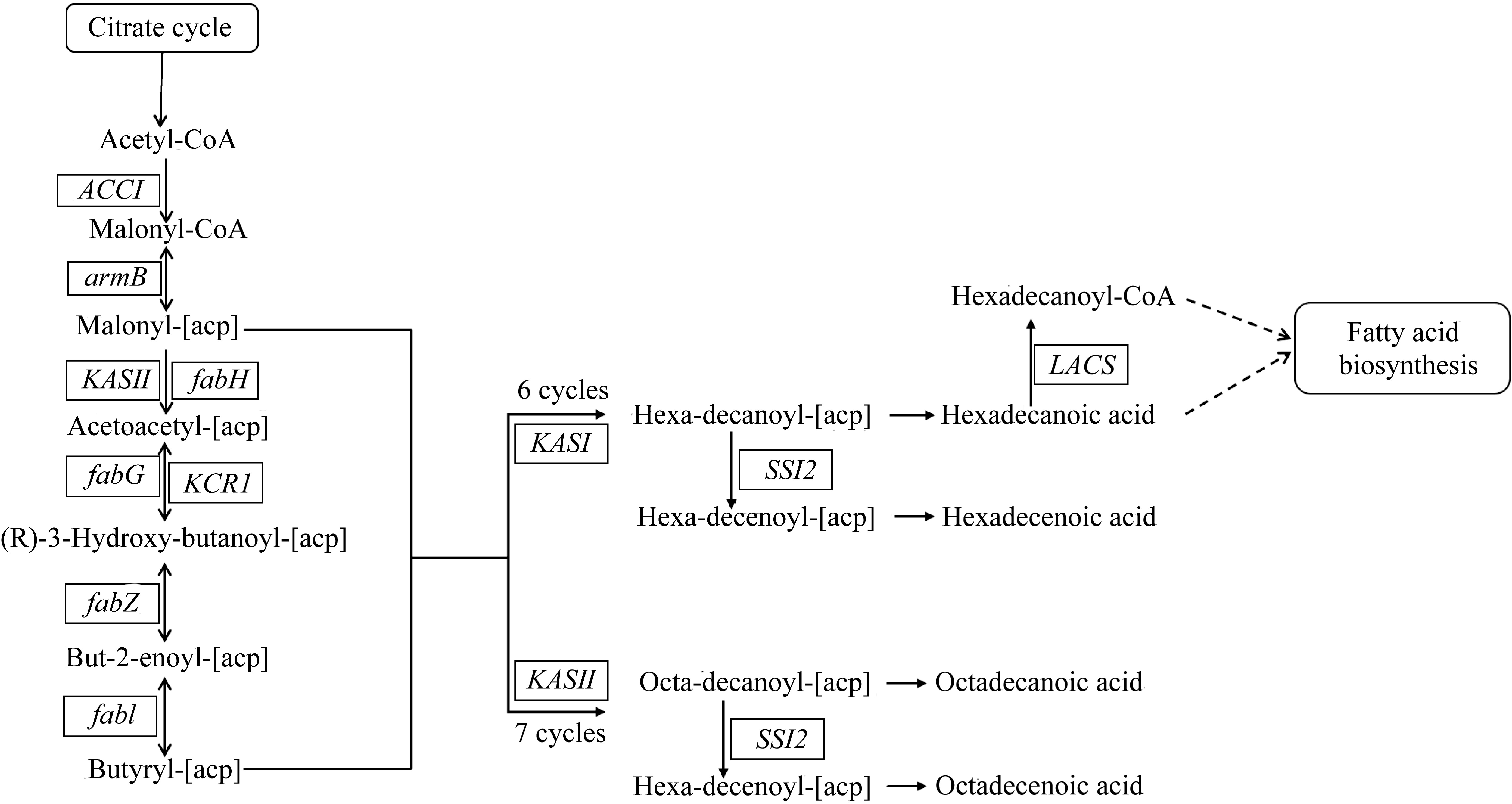
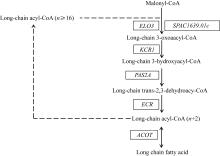
Tab. 1
Enzymes and transporters which is associated with fatty acid metabolism in the T. pseudonana transcriptome"
| 信号通路 | 酶(蛋白)名称 | EC编号 | 基因ID | 基因名称 |
|---|---|---|---|---|
| 脂肪酸的 生物合成 | 乙酰-CoA羧化酶(Acetyl-CoA carboxylase) | [EC:6.4.1.2] | THAPS_6770 | ACC1 |
| THAPSDRAFT_12234 | ACC1 | |||
| 丙二酸单酰CoA转移酶([acyl-carrier-protein] S-malonyltransferase) | [EC:2.3.1.39] | THAPSDRAFT_5219 | armB | |
| β-酮酰-ACP合酶Ⅱ (3-oxoacyl-[acyl-carrier-protein] synthase Ⅱ) | [EC:2.3.1.179] | THAPSDRAFT_961 | KASⅡ | |
| THAPSDRAFT_12152 | KAS | |||
| THAPSDRAFT_5021 | KASⅠ | |||
| THAPSDRAFT_262879 | KAS | |||
| β-酮酰-ACP合酶Ⅲ (3-oxoacyl-[acyl-carrier-protein] synthase Ⅲ) | [EC:1.1.1.180] | THAPSDRAFT_270141 | fabH | |
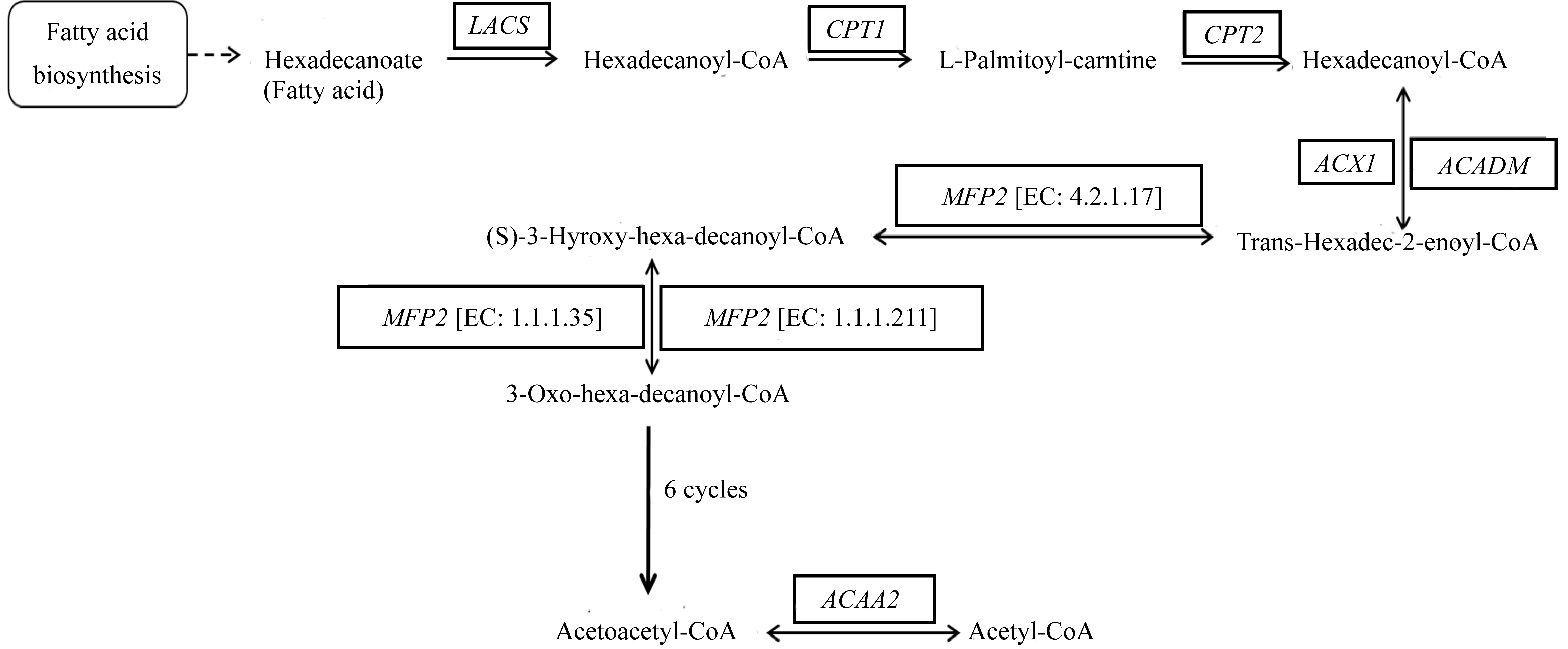
| 长链酰基-CoA合成酶(Long chain acyl-CoA synthetase) | [EC:6.2.1.3] | THAPS_263105 | AAE16 | |
| THAPSDRAFT_11953 | LACS7 | |||
| THAPSDRAFT_270334 | AAE15 | |||
| THAPSDRAFT_29867 | LACS7 | |||
| THAPSDRAFT_13156 | LACS8 | |||
| THAPSDRAFT_25042 | LACS7 | |||
| THAPSDRAFT_262242 | LACS6 | |||
| β-羟酰-ACP脱水酶(3-hydroxyacyl-[acyl-carrier-protein] dehydratase) | [EC:4.2.1.59] | THAPSDRAFT_34681 | fabZ | |
| 烯酰-ACP还原酶(Enoyl-[acyl-carrier protein] reductase I) | [EC:1.3.1.9] | THAPSDRAFT_32860 | fabI | |
| β-酮酰-ACP还原酶(3-oxoacyl-[acyl-carrier-protein] reductase) | [EC:1.1.1.100] | THAPSDRAFT_270321 | fabG | |
| 酰基-ACP去饱和酶(Stearoyl-[acyl-carrier-protein]9-desaturase 2) | [EC:1.14.19.2] | THAPSDRAFT_bd1192 | SSI2 | |
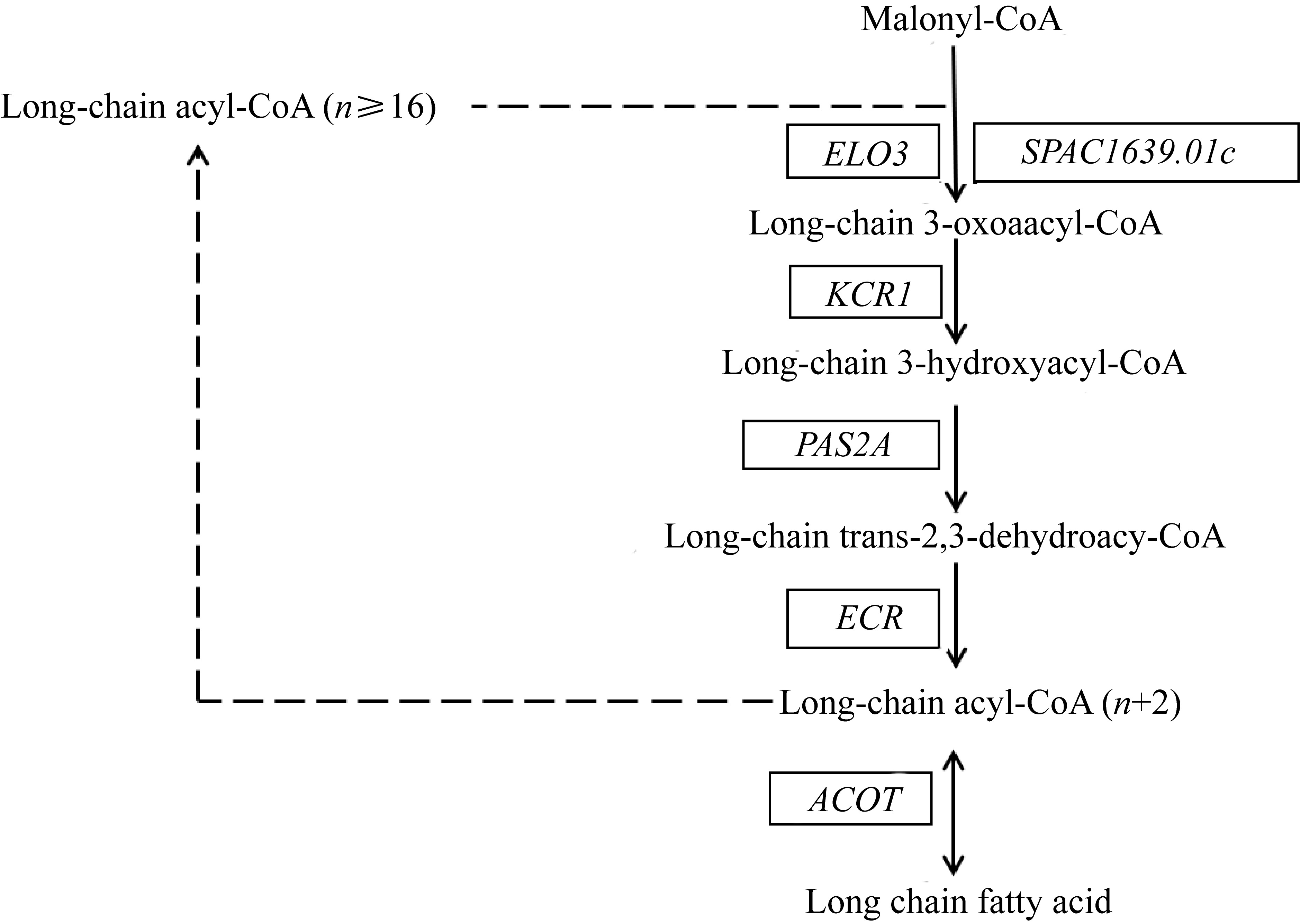
| 脂肪酸的 延伸 | 超长链脂肪酸延伸蛋白(Elongation of fatty acids protein) | [EC:2.3.1.199] | THAPSDRAFT_3741 | ELO3 |
| THAPSDRAFT_728 | SPAC1639.01c | |||
| THAPSDRAFT_22362 | KCS9 | |||
| 乙酰-CoA转移酶(Acetyl-CoA acetyltransferase) | [EC:2.3.1.16] | THAPSDRAFT_34809 | ACAA2 | |
| 超长链酮酰-CoA还原酶(Very-long-chain 3-oxoacyl-CoA reductase) | [EC:1.1.1.330] | THAPSDRAFT_22650 | KCR1 | |
| THAPS_35459 | ACLA_070510 | |||
| 超长链羟酰-CoA脱水酶(Very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase) | [EC:4.2.1.134] | THAPS_6781 | PAS2A | |
| 烯酰-CoA水合酶(Probable enoyl-CoA hydratase 1) | [EC:4.2.1.17] | THAPSDRAFT_34511 | ECHS1 |
| [1] |
曹月蕾, 2021. 集胞藻PCC6803类胡萝卜素和脂肪酸代谢途径关键基因功能研究[D]. 济南: 山东师范大学.
|
|
|
|
| [2] |
高彦龙, 吴玉霞, 张仲兴, 等, 2022. 苹果ELO家族基因鉴定及其在低温胁迫下的表达分析[J]. 园艺学报, 49(8): 1621-1636.
doi: 10.16420/j.issn.0513-353x.2021-0545 |
|
|
|
| [3] |
高羽荞, 闫博巍, 赵莹, 等, 2018. 碱蓬转录组分析及油脂合成相关基因表达模式[J]. 中国油料作物学报, 40(6): 801-811.
doi: 10.7505/j.issn.1007-9084.2018.06.009 |
|
|
|
| [4] |
黄璐, 2020. 海洋硅藻假微型海链藻休眠细胞形成过程的应激响应与长期适应机制[D]. 厦门: 厦门大学.
|
|
|
|
| [5] |
刘淑雅, 陈楠生, 2021. 胶州湾海域浮游植物和赤潮物种的生物多样性研究进展[J]. 海洋科学, 45(4): 170-188.
|
|
|
|
| [6] |
刘璇, 张莹, 李爱芬, 2022. 活性氧介导微藻脂质积累的研究现状及进展[J]. 植物生理学报, 58(7): 1201-1211.
|
|
|
|
| [7] |
聂煜东, 耿媛媛, 张贤明, 等, 2021. 产油微藻胁迫培养策略研究综述[J]. 中国环境科学, 41(8): 3853-3866.
|
|
|
|
| [8] |
王延莉, 曹阳, 梁祎凡, 等, 2022. 绵羊ACAA1基因多态性及其与肉质性状的关联性分析[J]. 吉林农业大学学报, 44(2): 204-209.
|
|
|
|
| [9] |
位文倩, 孙昕, 黄峰, 等, 2022. 转录组揭示自养和混合培养栅藻油脂代谢途径差异[J]. 中国油料作物学报, 44(1): 130-137.
doi: 10.19802/j.issn.1007-9084.2020325 |
|
|
|
| [10] |
于仁成, 吕颂辉, 齐雨藻, 等, 2020. 中国近海有害藻华研究现状与展望[J]. 海洋与湖沼, 51(4): 768-788.
|
|
|
|
| [11] |
张梅, 米铁柱, 甄毓, 等, 2018. 基于玛氏骨条藻转录组的脂肪酸合成途径分析[J]. 中国海洋大学学报, 48(4): 81-93.
|
|
|
|
| [12] |
周婧雯, 丁玉娇, 曾晓蓉, 等, 2020. 生菜种子低温处理后耐冻性和脂肪酸合成代谢的关系研究[J]. 亚热带植物科学, 49(1): 9-14.
|
|
|
|
| [13] |
庄珊珊, 2019. 海洋硅藻假微型海链藻休眠细胞形成过程的相关代谢调控[D]. 厦门: 厦门大学.
|
|
|
|
| [14] |
doi: 10.1016/j.biortech.2019.121514 |
| [15] |
doi: 10.1038/srep11213 |
| [16] |
doi: 10.1016/j.biortech.2016.07.022 |
| [17] |
doi: 10.1016/j.cub.2016.05.056 |
| [18] |
doi: 10.1093/bioinformatics/btu170 pmid: 24695404 |
| [19] |
doi: 10.1007/s11274-013-1253-0 |
| [20] |
doi: 10.1126/science.281.5374.237 |
| [21] |
|
| [22] |
|
| [23] |
doi: 10.3390/biology10070565 |
| [24] |
doi: 10.1016/j.phytochem.2011.12.007 |
| [25] |
doi: 10.1111/j.0022-3646.1996.00479.x |
| [26] |
|
| [27] |
doi: 10.1093/plankt/fbu029 |
| [28] |
doi: 10.1016/S0021-9258(19)36507-X |
| [29] |
doi: 10.1016/j.algal.2015.09.006 |
| [30] |
doi: 10.1016/j.bbalip.2018.10.004 |
| [31] |
doi: 10.1104/pp.104.054528 |
| [32] |
doi: 10.1016/j.apenergy.2010.11.029 |
| [1] | Xiaolan PAN, Huiru LIU, Meng XU, Hanzhi XU, Hua ZHANG, Maoxian HE. Cloning and expression analysis of aquaporin gene AQP4 cDNA from Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2020, 39(3): 66-75. |
| [2] | Fuxuan WANG, Shu XIAO, Zhiming XIANG, Ziniu YU. Molecular cloning and expression analysis of Commd1 under salinity stress in Crassostrea hongkongensis [J]. Journal of Tropical Oceanography, 2017, 36(1): 48-55. |
| [3] | YANG Yu-qing,YU De-guang,XIE Jun,YU Er-meng,LI Wang-dong,WANG Guang-. Effects of acute salinity stress on Na+/K+-ATPase activity and plasma indicators of Epinephelus coioides [J]. Journal of Tropical Oceanography, 2010, 29(4): 160-164. |
|
||