Journal of Tropical Oceanography ›› 2018, Vol. 37 ›› Issue (1): 1-11.doi: 10.11978/2017041CSTR: 32234.14.2017041
• Orginal Article • Next Articles
Characteristic analysis of the complete mitogenome of Plagiopsetta glossa and a possible mechanism for gene rearrangement
Jiangxing DONG1,2( ), Wei SHI1, Xiaoyu KONG1(
), Wei SHI1, Xiaoyu KONG1( ), Shixi CHEN1,2
), Shixi CHEN1,2
- 1. CAS Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Guangzhou 510301, China
2. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2017-04-11Revised:2017-05-25Online:2018-01-20Published:2018-02-02 -
Supported by:National Natural Science Foundation of China (31471979, 30870283)
CLC Number:
- P735.541
Cite this article
Jiangxing DONG, Wei SHI, Xiaoyu KONG, Shixi CHEN. Characteristic analysis of the complete mitogenome of Plagiopsetta glossa and a possible mechanism for gene rearrangement[J].Journal of Tropical Oceanography, 2018, 37(1): 1-11.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
The primers used for amplification in Plagiopsetta glossa mitogenomes"
| 正向引物 | 序列(5′—3′) | 反向引物 | 序列(5′—3′) |
|---|---|---|---|
| Z15 | ATTAAAGCATAACHCTGAAGATGTTAAGAT | F2753 | TAGATAGAAACTGACCTGGATTACTCCGGT |
| Z2625 | GTTTACGACCTCGATGTTGGATCAGGACAT | F6746 | GCGGTGGATTGTAGACCCATARACAGAGGT |
| Z-Ile | AAGGRHYACTTTGATAGAG | FND5-1180 | ATRATNGCRTCYTTDGARWARAANCC |
| ZND5-1050 | GCNATRCTNTTYYTNTGYTCNGGNTC | F-Cytb | TGNCCRATRAKVAYRWADGGRTSTTC |
| ZND6-210 | GCHARNGCNRCHGARTANGCAAA | FCOI-70 | CCHACYATNCCDGCYCARGCMCCRAA |
| Z6468 | CCACATCTDCTGCATGCAAAYCAYACACTT | FATP6-495 | AGRTGNCCNGCNGTBARRTTNGCNGT |
| ZATP-310 | ACHTTYACNCCHACHACNCARCTNTC | FND4-865 | CCYATGTGVCYNACDGADGAGTADGC |
| Z10818 | TTYGAAGCAGCCGCMTGATACTGACAYTT | F13413 | TAGCTGCTACTCGGATTTGCACCAAGAGT |
| Z13347 | AAGGATAACAGCTCATCCGTTGGTCTTAGG | F49 | GGCCCATCTTAACATCTTC |
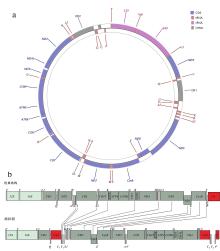
Fig. 1
Gene map of P. glossa mitogenomeand the features of gene rearrangement. (a) Gene map of P. glossa mitogenome; (b) the features of gene rearrangement. repeat region representes repeat sequence. Protein-coding genes are indicated by dark green boxes, rRNA by light green boxes, CRs (control region) and NC (non-coding region) by red boxes, and tRNA by white columns. The genes and their full names are listed in Tab. 2. Genes labeled above the diagram are encoded by the H-strand, and the others by the L-strand"
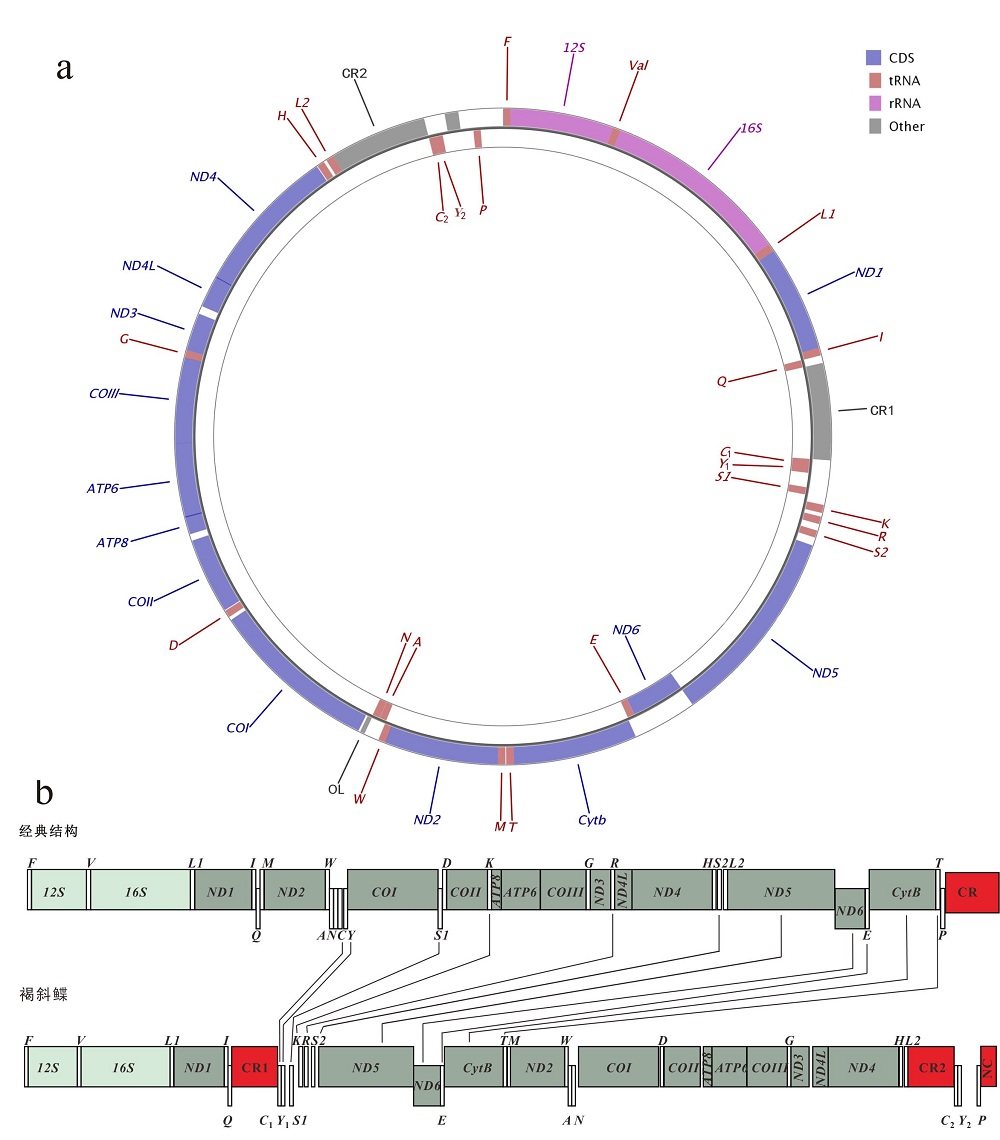
Tab. 2
Organization of P. glossa mitochondrial genome"
| 基因 | 位置 | 长度 | 密码子 | 间隔区 | |||||
|---|---|---|---|---|---|---|---|---|---|
| 起始 | 终止 | bp | aa | 起始 | 终止 | 反密码子 | 编码链 | ||
| Phe(F) | 1 | 68 | 68 | GAA | H | 0 | |||
| 12S | 69 | 1019 | 951 | H | 0 | ||||
| Val(V) | 1020 | 1091 | 72 | TAC | H | 0 | |||
| 16S | 1092 | 2814 | 1723 | H | 0 | ||||
| Leu1(L1) | 2815 | 2888 | 74 | TAA | H | 0 | |||
| ND1 | 2889 | 3863 | 975 | 324 | ATG | TAA | H | 0 | |
| Ile(I) | 3864 | 3935 | 72 | GAT | H | 0 | |||
| Gln(Q) | 3934 | 4005 | 72 | TTG | L | -2 | |||
| CR1 | 4006 | 4898 | 893 | H | 0 | ||||
| Cys1(C1) | 4899 | 4966 | 68 | GCA | L | 0 | |||
| Tyr1(Y1) | 4967 | 5034 | 68 | GTA | L | 0 | |||
| Ser1(S1) | 5176 | 5246 | 71 | TGA | L | 141 | |||
| Lys(K) | 5309 | 5383 | 75 | TTT | H | 62 | |||
| Arg(R) | 5418 | 5487 | 70 | TCG | H | 34 | |||
| Ser2(S2) | 5549 | 5615 | 67 | GCT | H | 61 | |||
| ND5 | 5691 | 7529 | 1839 | 612 | ATG | TAA | H | 75 | |
| ND6 | 7526 | 8047 | 522 | 173 | ATG | TAG | L | -4 | |
| Glu(E) | 8049 | 8117 | 69 | TTC | L | 1 | |||
| Cytb | 8122 | 9262 | 1141 | 380 | ATG | T | H | 4 | |
| Thr(T) | 9263 | 9333 | 71 | TGT | H | 0 | |||
| Met(M) | 9341 | 9409 | 69 | CAT | H | 7 | |||
| ND2 | 9410 | 10456 | 1047 | 348 | ATG | TAG | H | 0 | |
| Trp(W) | 10455 | 10524 | 70 | TCA | H | -2 | |||
| Ala(A) | 10526 | 10593 | 68 | TGC | L | 1 | |||
| Asn(N) | 10595 | 10667 | 73 | GTT | L | 1 | |||
| OL | 10663 | 10706 | 44 | L | -5 | ||||
| COⅠ | 10724 | 12274 | 1551 | 516 | GTG | TAA | H | 17 | |
| Asp(D) | 12306 | 12373 | 68 | GTC | H | 31 | |||
| COⅡ | 12381 | 13079 | 699 | 232 | ATG | AGA | H | 7 | |
| ATP8 | 13141 | 13308 | 168 | 55 | ATG | TAA | H | 61 | |
| ATP6 | 13299 | 13982 | 684 | 227 | ATG | TAA | H | -10 | |
| COⅢ | 13982 | 14767 | 786 | 261 | ATG | TAA | H | -1 | |
| Gly(G) | 14767 | 14836 | 70 | TCC | H | -1 | |||
| ND3 | 14837 | 15187 | 351 | 116 | ATG | TAA | H | 0 | |
| ND4L | 15261 | 15557 | 297 | 98 | ATG | TAA | H | 73 | |
| ND4 | 15551 | 16924 | 1374 | 457 | ATG | AGA | H | -7 | |
| His(H) | 16932 | 17000 | 69 | GTG | H | 7 | |||
| Leu2(L2) | 17028 | 17100 | 73 | TAG | H | 27 | |||
| CR2 | 17101 | 17997 | 897 | H | 0 | ||||
| Cys2(C2) | 17998 | 18065 | 68 | GCA | L | 0 | |||
| Tyr2(Y2) | 18066 | 18133 | 68 | GTA | L | 0 | |||
| Pro(P) | 18436 | 18506 | 71 | TGG | L | 302 | |||
| NC | 18507 | 18723 | 217 | H | 0 | ||||
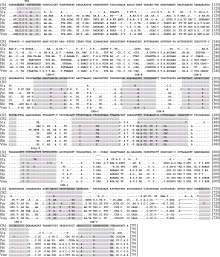
Fig. 2
Aligned sequences of the control regions of P. glossa and seven other flatfish. CR1 and CR2: the two control regions in P. glossa mitogenome; Sla: Samariscus latus, NC_024263; Pol: Paralichthys olivaceus, NC_002386; Pco: Pleuronichthys cornutus NC_022445; Hhi: Hippoglossus hippoglossus, NC_009709; Hst: Hippoglossus stenolepis, NC_009710; Rhi: Reinhardtius hippoglossoides, NC_009711; Vmo: Verasper moseri, NC_008461. The shaded sequences represent the conservative blocks. TAS: terminal associated sequence; CSB: conserved sequence block"


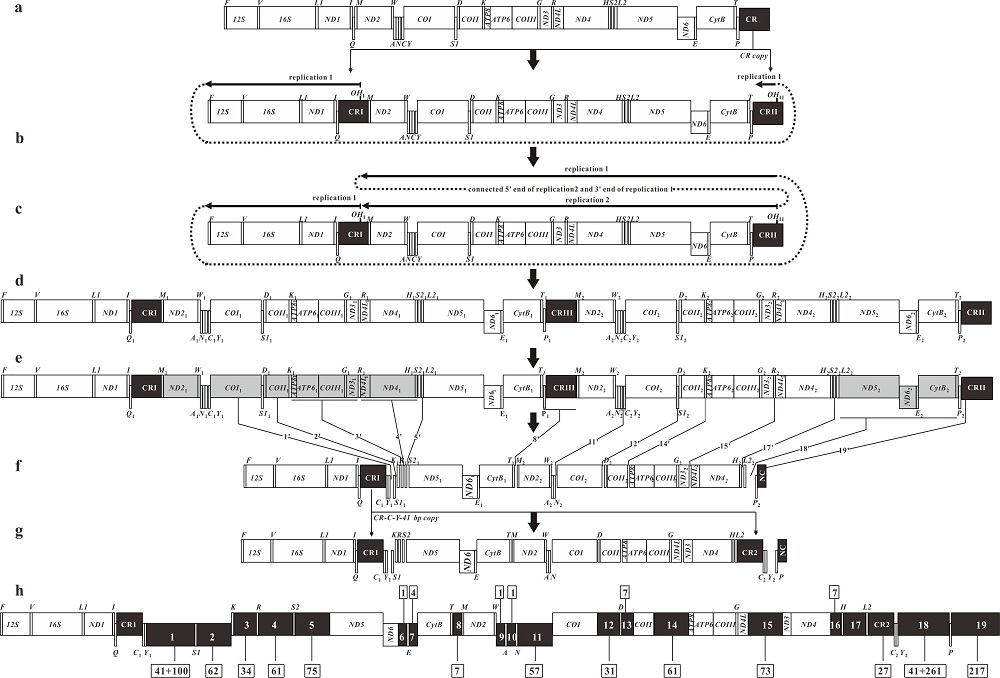
Fig. 3
The inferred mechanism of gene rearrangement from the ancestral gene order to that of P. glossa mitogenome based on the DRRL model. Protein-coding genes and rRNA are indicated by boxes, and tRNA genes are indicated by columns. Genes labeled above the diagram are encoded by the H-strand, and the others, by the L-strand. OH indicates the origin of replication for the H-strand; the direction of replication is shown by arrows. The dark boxes indicate the control regions (CRs), noncoding region (NC) and intergenic spacers. (a) The ancestral mitogenome with 37 genes and one CR. (b) The ancestral CR was duplicated to CRⅠ and CRⅡ, and then CRⅠ was translocated to the position between tRNA-Q and tRNA-M. A first mitochondrial replication event (RP1) was initiated at OHⅠ. (c) After RP1 passed through OHⅡ, secondary replication (RP2) began at OHⅡ. Both replications terminated close to the site of OHⅠ. (d) The duplication of 29 genes and one CR yielded by double replications. (e) One of each copied gene pair was lost randomly. Dark gray boxes indicate degenerated genes. The italic lines indicate the positions of intergenetic spaces from the degenerated genes. The numbers marked by prime indicate the order of the intergenetic spaces in genenome. (f) The order of genes, CRs and intergenic spaces after the loss of several genes. (g) The mitogenome of P. glossa was formed after CRⅠ-tRNA-C1-Y1 and the first 41 bp of Non-coding sequence 1 duplicated and translocated to the position between tRNA-L2 and tRNA-P. (h) The positions and lengths of intergenic spacers of P. glossa. The number in black shaded boxes indicate the intergenic spacer order in mitogenome. The numbers in the boxes above and below represent the length of the intergenic spacers. The boxes above the diagram indicate the sites of intergenic spacers same as the typical one in teleosts; the others are only in P. glossa"
| [1] | 龚理, 时伟, 司李真, 等, 2013. 鱼类线粒体DNA重排研究进展[J]. 动物学研究, 34(6): 666-673. |
| GONG LI, SHI WEI, SI LIZHEN, et al, 2013. Rearrangement of mitochondrial genome in fishes[J]. Zoological Research, 34(6): 666-673 (in Chinese with English abstract). | |
| [2] | 刘焕章, 2002. 鱼类线粒体DNA控制区的结构和进化: 以鳑鲏鱼类为例[J]. 自然科学进展, 12(3): 266-270. |
| LIU HUANZHANG, 2002. The structure and evolution of the mtDNA control region in fish: taking example for Acheilognathinae[J]. Progress in Natural Science, 12(3): 266-270 (in Chinese). | |
| [3] | ARNDT A, SMITH M J, 1998. Mitochondrial gene rearrangement in the sea cucumber genus Cucumaria[J]. Molecular Biology and Evolution, 15(8): 1009-1016. |
| [4] | BOORE J L, 1999. Animal mitochondrial genomes[J]. Nucleic Acids Research, 27(8): 1767-1780. |
| [5] | CANTATORE P, GADALETA M N, ROBERTI M, et al, 1987. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes[J]. Nature, 329(6142): 853-855. |
| [6] | CHEN PENGYAN, ZHENG BOYING, LIU JINGXIAN, et al, 2016. Next-generation sequencing of two mitochondrial genomes from family pompilidae (Hymenoptera: Vespoidea) reveal novel patterns of gene arrangement[J]. International Journal of Molecular Sciences, 17(10): 1641. |
| [7] | GONG LI, SHI WEI, YANG MIN, et al, 2016. Novel gene arrangement in the mitochondrial genome of Bothus myriaster (Pleuronectiformes: Bothidae): evidence for the Dimer- Mitogenome and Non-random Loss model[J]. Mitochondrial DNA Part A, 27(5): 3089-3092. |
| [8] | GUO AIJIANG, 2016. The complete mitochondrial genome of the tapeworm Cladotaenia vulturi (Cestoda: Paruterinidae): gene arrangement and phylogenetic relationships with other cestodes[J]. Parasites & Vectors, 9(1): 475. |
| [9] | HALL T A, 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT[J]. Nucleic Acids Symposium Series, 41(2): 95-98. |
| [10] | INOUE J G, MIYA M, TSUKAMOTO K, et al, 2003. Evolution of the deep-sea gulper eel mitochondrial genomes: large-scale gene rearrangements originated within the eels[J]. Molecular Biology and Evolution, 20(11): 1917-1924. |
| [11] | JACOBS H T, HERBERT E R, RANKINE J, 1989. Sea urchin egg mitochondrial DNA contains a short displacement loop (D-loop) in the replication origin region[J]. Nucleic Acids Research, 17(22): 8949-8965. |
| [12] | KONG XIAOYU, DONG XIAOLI, ZHANG YANCHUN, et al, 2009. A novel rearrangement in the mitochondrial genome of tongue sole, Cynoglossus semilaevis: control region translocation and a tRNA gene inversion[J]. Genome, 52(12): 975-984. |
| [13] | LAVROV D V, BOORE J L, BROWN W M, 2002, Complete mtDNA sequences of two millipedes suggest a new model for mitochondrial gene rearrangements: duplication and nonrandom loss[J]. Molecular Biology and Evolution, 19(2): 163-169. |
| [14] | LEE J, LEE H, LEE J, et al, 2015. Complete mitochondrial genome of the Antarctic silverfish, Pleuragramma antarcticum (Perciformes, Nototheniidae)[J]. Mitochondrial DNA, 26(6): 885-886. |
| [15] | LEE J S, MIYA M, LEE Y S, et al, 2001. The complete DNA sequence of the mitochondrial genome of the self-fertilizing fish Rivulus marmoratus (Cyprinodontiformes, Rivulidae) and the first description of duplication of a control region in fish[J]. Gene, 280(1/2): 1-7. |
| [16] | LI DONGHE, SHI WEI, GONG LI, et al, 2016. The complete mitochondrial genome of Paraplagusia blochii (Pleuronectiformes: Cynoglossidae)[J]. Mitochondrial DNA Part A, 27(1): 92-93. |
| [17] | LI YUAN, SONG NA, LIN LONGSHAN, et al, 2016. The complete mitochondrial genome of Pampus echinogaster (Perciformes: Stromateidae)[J]. Mitochondrial DNA Part A, 27(1): 289-290. |
| [18] | LIN CHUYIN, LIN WENWEN, KAO H W, 2012. The complete mitochondrial genome of the mackerel icefish, Champsocephalus gunnari (Actinopterygii: Channichthyidae), with reference to the evolution of mitochondrial genomes in Antarctic notothenioids[J]. Zoological Journal of the Linnean Society, 165(3): 521-533. |
| [19] | LOWE T M, EDDY S R, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence[J]. Nucleic Acids Research, 25(5): 955-964. |
| [20] | LUNT D H, HYMAN B C, 1997. Animal mitochondrial DNA recombination[J]. Nature, 387(6630): 247. |
| [21] | MORITZ C, BROWN W M, 1987. Tandem duplications in animal mitochondrial DNAS: variation in incidence and gene content among lizards[J]. Proceedings of the National Academy of Sciences of the United States of America, 84(20): 7183-7187. |
| [22] | MU XIDONG, YANG YEXIN, LIU YI, et al, 2017. The complete mitochondrial genomes of two freshwater snails provide new protein-coding gene rearrangement models and phylogenetic implications[J]. Parasites & Vectors, 10: 11. |
| [23] | NELSON J S, 2006. Fishes of the word[M]. 4th ed. Portland, OR: John Wiley & Sons, Inc.: 442-451. |
| [24] | OH J S, KANG S, AHN D H, et al, 2016. Complete mitochondrial genome of the Antarctic dragonfish, Parachaenichthys charcoti (Notothenioidei: Bathydraconidae)[J]. Mitochondrial DNA Part A, 27(5): 3151-3152. |
| [25] | POULTON J, DEADMAN M E, BINDOFF L, et al, 1993. Families of mtDNA-rearrangements can be detected in patients with mtDNA deletions: duplications may be a transient intermediate form[J]. Human Molecular Genetics, 2(1): 23-30. |
| [26] | SATOH T P, MIYA M, ENDO H, et al, 2006. Round and pointed-head grenadier fishes (Actinopterygii: Gadiformes) represent a single sister group: evidence from the complete mitochondrial genome sequences[J]. Molecular Phylogenetics and Evolution, 40(1): 129-138. |
| [27] | SHADEL G S, CLAYTON D A, 1997. Mitochondrial DNA maintenance in vertebrates[J]. Annual Review of Biochemistry, 66: 409-435. |
| [28] | SHI WEI, DONG XIAOLI, WANG ZHONHMING, et al, 2013. Complete mitogenome sequences of four flatfishes (Pleuronectiformes) reveal a novel gene arrangement of L-strand coding genes[J]. BMC Evolutionary Biology, 13: 173. |
| [29] | SHI WEI, MIAO XIANGUANG, KONG XIAOYU, 2014. A novel model of double replications and random loss accounts for rearrangements in the Mitogenome of Samariscus latus (Teleostei: Pleuronectiformes)[J]. BMC Genomics, 15: 352. |
| [30] | SHI WEI, GONG LI, WANG SHUYING, et al, 2015. Tandem Duplication and Random Loss for mitogenome rearrangement in Symphurus (Teleost: Pleuronectiformes)[J]. BMC Genomics, 16: 355. |
| [31] | SHI WEI, GONG LI, KONG XIAOYU, 2016. The complete mitochondrial genome sequence of Cynoglossus abbreviatus (Pleuronectiformes: Cynoglossidae) with control region translocation and tRNA-Gln gene inversion[J]. Mitochondrial DNA Part A, 27(3): 2157-2158. |
| [32] | STOTHARD P, WISHART D S, 2005. Circular genome visualization and exploration using CGView[J]. Bioinformatics, 21(4): 537-539. |
| [33] | THOMPSON J D, GIBSON T J, PLEWNIAK F, et al, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools[J]. Nucleic Acids Research, 25(24): 4876-4882. |
| [34] | VERKUIL Y I, PIERSMA T, BAKER A J, 2010. A novel mitochondrial gene order in shorebirds (Scolopacidae, Charadriiformes)[J]. Molecular Phylogenetics and Evolution, 57(1): 411-416. |
| [35] | WANG SHUYING, SHI WEI, MIAO XIANGUANG, et al, 2014. Complete mitochondrial genome sequences of three rhombosoleid fishes and comparative analyses with other flatfishes (Pleuronectiformes)[J]. Zoological Studies, 53: 80. |
| [36] | XIA YUN, ZHENG YUCHI, MURPHY R W, et al, 2016. Intraspecific rearrangement of mitochondrial genome suggests the prevalence of the tandem duplication-random loss (TDLR) mechanism in Quasipaa boulengeri[J]. BMC Genomics, 17: 965. |
| [37] | YANG MIN, SHI WEI, MIAO XIANGUANG, et al, 2016. The complete mitochondrial genome of Cynoglossus puncticeps (Pleuronectiformes: Cynoglossidae)[J]. Mitochondrial DNA Part A, 27(2): 1233-1234. |
| [38] | ZHUANG XUAN, QU MENG, ZHANG XIANG, et al, 2013. A comprehensive description and evolutionary analysis of 22 grouper (Perciformes, Epinephelidae) mitochondrial genomes with emphasis on two novel genome organizations[J]. PLoS One, 8(8): e73561. |
| [1] | HUANG Peixian, YAO Xuemei, YU Qiaochi, ZHANG Jiayu. Mitogenome characteristics and phylogenetic analysis of Siphonosoma australe in Hainan [J]. Journal of Tropical Oceanography, 2023, 42(2): 54-63. |
| [2] | ZHANG Shaoqiu, KONG Xiaoyu. A new record genus—Taeniopsetta and species—Taeniopsetta ocellata of Bothidae from the coastal waters of mainland China [J]. Journal of Tropical Oceanography, 2022, 41(1): 1-6. |
| [3] | MIAO Xian-guang, JIANG Jin-xia, SHI Wei, WANG Zhong-ming, WANG Shu-ying, KONG Xiao-yu. Phylogenetic relationship of Cynoglossinae based on COI barcoding marker [J]. Journal of Tropical Oceanography, 2013, 32(5): 85-92. |
| [4] | ZHANG Yan-chun,KONG Xiao-yu,WANG Zhong-ming. Characterization of mitochondrial control region sequence of Psettodes erumei and phylogenetic analysis of Pleuronectiformes [J]. Journal of Tropical Oceanography, 2010, 29(6): 71-78. |
|
||

