Journal of Tropical Oceanography ›› 2021, Vol. 40 ›› Issue (1): 65-74.doi: 10.11978/2020024CSTR: 32234.14.2020024
• Marine Biology • Previous Articles Next Articles
Cloning and functional analysis of a new pattern recognition receptor CgLRRC69 in Pacific oyster, Crassostrea gigas
ZHANG Xiangyu1,3( ), SONG Jingchen2, LIU Kunna1,3, MAO Fan1, XIAO Shu1, XIANG Zhiming1, ZHANG Yang1(
), SONG Jingchen2, LIU Kunna1,3, MAO Fan1, XIAO Shu1, XIANG Zhiming1, ZHANG Yang1( ), YU Ziniu1(
), YU Ziniu1( )
)
- 1. Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Science, Guangzhou 510301, China
2. College of Oceanology, South China Agricultural University, Guangzhou 510642, China
3. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2020-02-28Revised:2020-04-28Online:2021-01-10Published:2020-05-19 -
Contact:ZHANG Yang,YU Ziniu E-mail:loginlv@163.com;yzhang@scsio.ac.cn;carlzyu@scsio.ac.cn -
Supported by:National Natural Science Foundation of China(31572640);National Natural Science Foundation of China(31572661)
CLC Number:
- P735.2
Cite this article
ZHANG Xiangyu, SONG Jingchen, LIU Kunna, MAO Fan, XIAO Shu, XIANG Zhiming, ZHANG Yang, YU Ziniu. Cloning and functional analysis of a new pattern recognition receptor CgLRRC69 in Pacific oyster, Crassostrea gigas[J].Journal of Tropical Oceanography, 2021, 40(1): 65-74.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Sequences of oligonucleotide primers used"
| 引物 | 序列 | 用途 | |||
|---|---|---|---|---|---|
| DsGFP-F | TAATACGACTCACTATAGGNGTCAGTGGAGAGGGTGAAGG | RNAi | |||
| DsGFP-R | TAATACGACTCACTATAGGNAGGCATGGCACTCTTGAAAA | RNAi | |||
| DsLRR-F | TAATACGACTCACTATAGGNTCCGCTTGAGTTTGGATTCC | RNAi | |||
| DsLRR-R | TAATACGACTCACTATAGGNTTCCTTCAGTGAGAGGACCT | RNAi | |||
| LRRi-F | AGAACTACCCGAAGTCCTTGA | qRT-PCR | |||
| LRRi-R | CCAAACTCAAGCGGAAGACT | qRT-PCR | |||
| GAPDH-F | CTGCCAACGTGTCAGTGGTG | qRT-PCR | |||
| GAPDH-R | TCAGTGTAGCCCAGGATGCC | qRT-PCR | |||
| LRR-F | AGCATTGATAGTTGAGATCA | ORF | |||
| LRR-R | TCGAAGAGCAAATCACATGA | ORF | |||

Fig. 1
The complete ORF and deduced amino acid sequence of CgLRRC69. The black font is the open reading frame (ORF) of Oyster CgLRRC69, the blue font is the translated amino acid sequence, and the bold font indicates the start codon and the stop codon. * represents the stop codon, which does not encode any amino acid and is responsible for terminating the peptide chain synthesis. Shading indicates the LRR-leucine repeat sequence"


Fig. 1
The complete ORF and deduced amino acid sequence of CgLRRC69. The black font is the open reading frame (ORF) of Oyster CgLRRC69, the blue font is the translated amino acid sequence, and the bold font indicates the start codon and the stop codon. * represents the stop codon, which does not encode any amino acid and is responsible for terminating the peptide chain synthesis. Shading indicates the LRR-leucine repeat sequence"

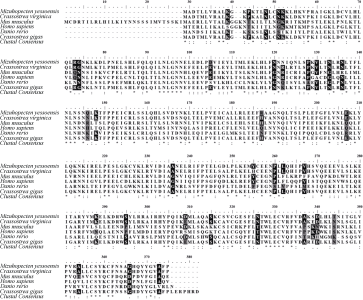
Fig. 2
The amino acid sequence of Pacific oyster LRRC69, compared with Homo sapiens, Mus musculus, Danio rerio, Mizuhopecten yessoensis, and Crassostrea virginica. Identical, highly conserved and less conserved amino acid residues are indicated by *, : and . . White letters on black background indicate that in the six selected species, there are still more than three identical amino acid residues in the less conservative amino acid residues. Black letters on gray background indicate that there are amino acids with similar properties in the less conservative amino acid residues. The GenBank accession numbers are listed: Homo sapiens (NP_001123362.1), Mus musculus (XP_006538358.1), Danio rerio (XP_005159720.1), Mizuhopecten yessoensis (XP_021346950.1) and Crassostrea virginica (XP_022338017.1)"


Fig. 2
The amino acid sequence of Pacific oyster LRRC69, compared with Homo sapiens, Mus musculus, Danio rerio, Mizuhopecten yessoensis, and Crassostrea virginica. Identical, highly conserved and less conserved amino acid residues are indicated by *, : and . . White letters on black background indicate that in the six selected species, there are still more than three identical amino acid residues in the less conservative amino acid residues. Black letters on gray background indicate that there are amino acids with similar properties in the less conservative amino acid residues. The GenBank accession numbers are listed: Homo sapiens (NP_001123362.1), Mus musculus (XP_006538358.1), Danio rerio (XP_005159720.1), Mizuhopecten yessoensis (XP_021346950.1) and Crassostrea virginica (XP_022338017.1)"

Fig. 3
Schematic of LRRC69 structures in different species. P means Pfam: LRR; T means LRR-TYP; blank means ordinary LRR. The GenBank accession numbers are listed: Homo sapiens (NP_001123362.1), Mus musculus (XP_006538358.1), Danio rerio (XP_005159720.1), Mizuhopecten yessoensis (XP_021346950.1) and Crassostrea virginica (XP_022338017.1)"
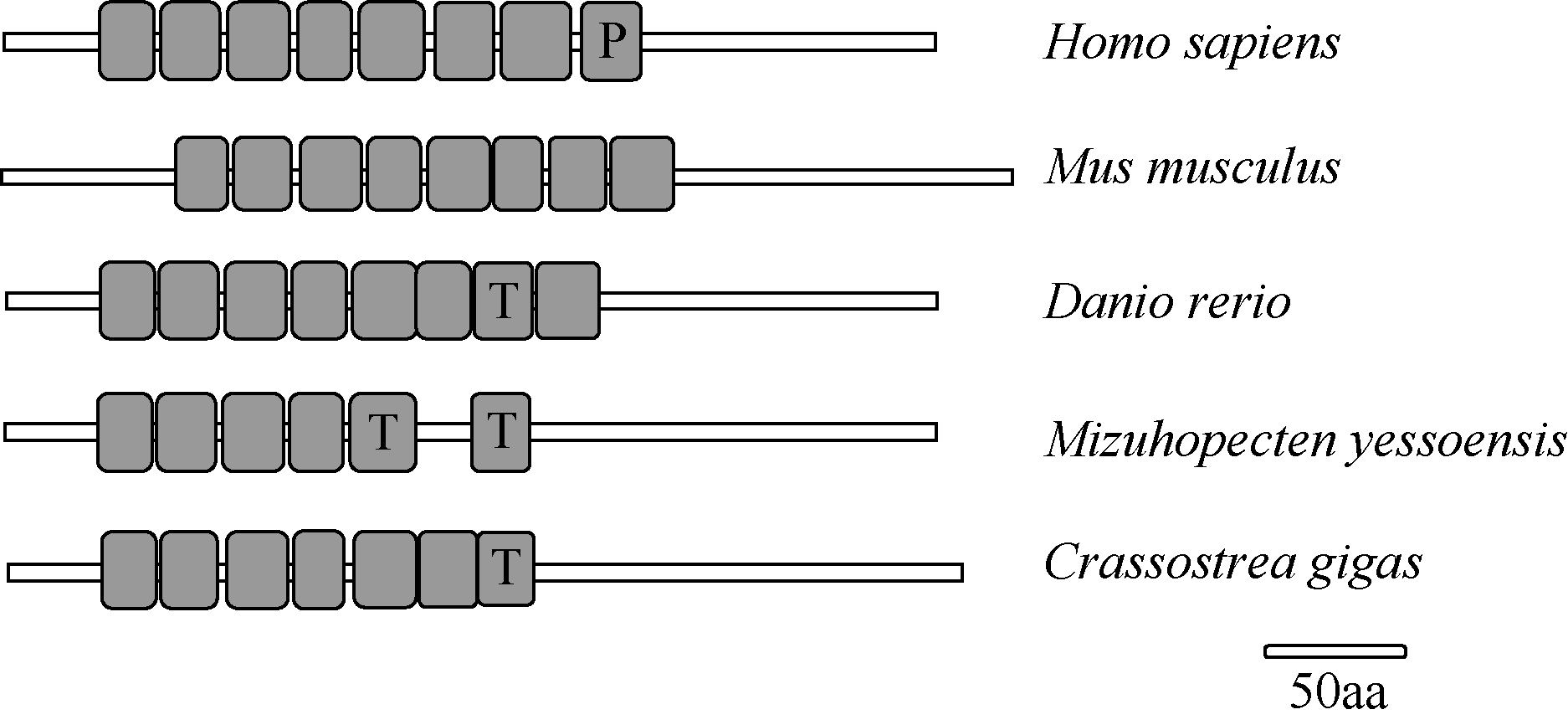

Fig. 3
Schematic of LRRC69 structures in different species. P means Pfam: LRR; T means LRR-TYP; blank means ordinary LRR. The GenBank accession numbers are listed: Homo sapiens (NP_001123362.1), Mus musculus (XP_006538358.1), Danio rerio (XP_005159720.1), Mizuhopecten yessoensis (XP_021346950.1) and Crassostrea virginica (XP_022338017.1)"

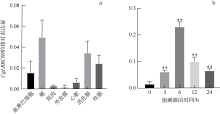
Fig. 5
CgLRRC69 gene expression in tissues and under bacterial challenge. (a) Tissue distribution of CgLRRC69 in oyster, including hemocytes, gill, muscle, mantle, heart, digestive gland, and gonads. (b) Variation in expression of CgLRRC69 in response to bacterial challenge. Data are expressed as Mean ± SEM (N=3). ** means p<0.01, namely, the difference is extremely significant"
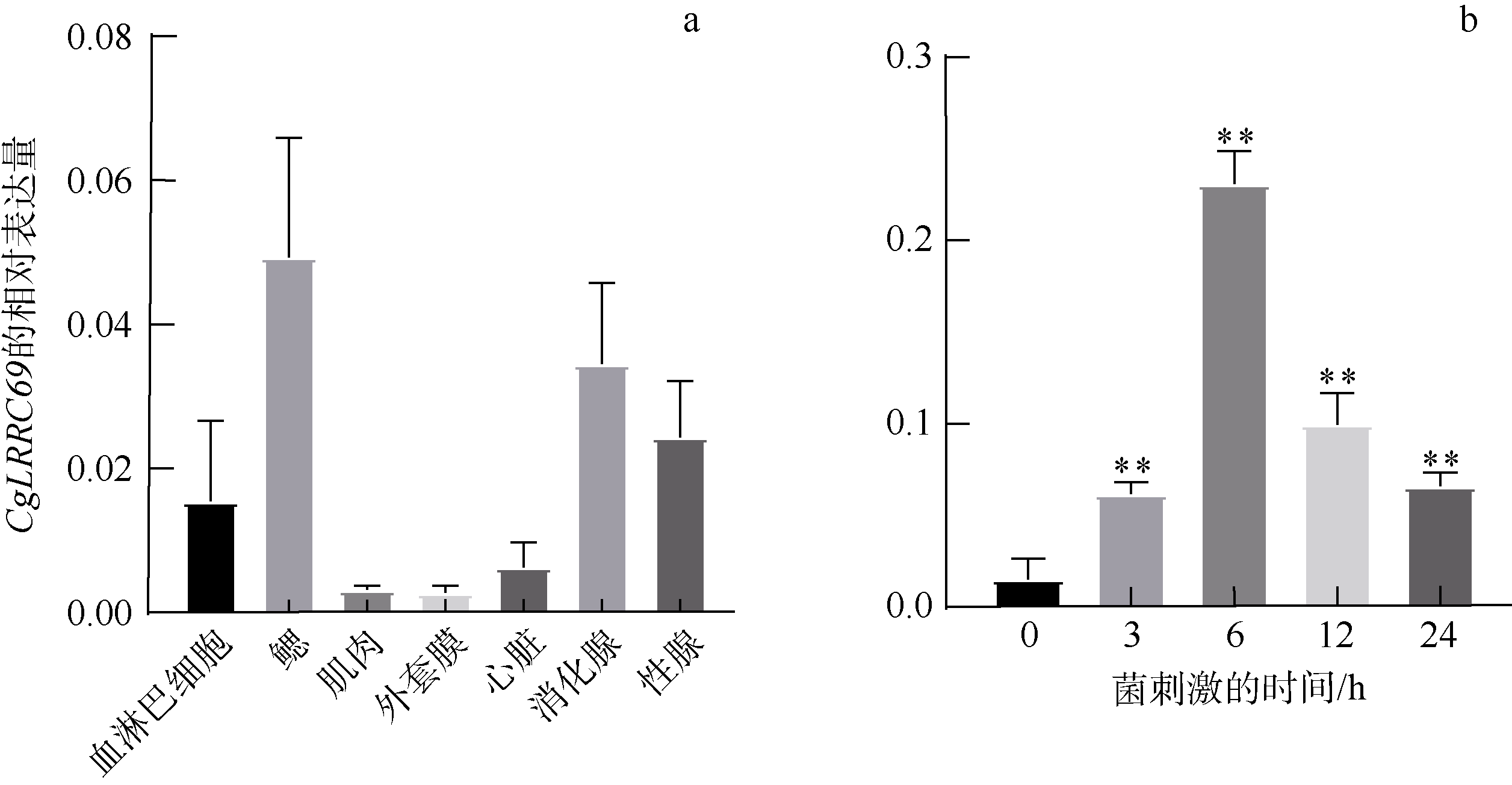

Fig. 5
CgLRRC69 gene expression in tissues and under bacterial challenge. (a) Tissue distribution of CgLRRC69 in oyster, including hemocytes, gill, muscle, mantle, heart, digestive gland, and gonads. (b) Variation in expression of CgLRRC69 in response to bacterial challenge. Data are expressed as Mean ± SEM (N=3). ** means p<0.01, namely, the difference is extremely significant"


Fig. 6
CgLRRC69 protein with affinity specificity for LPS. (a) The puri?ed CgLRRC69 recombinant protein was analyzed by SDS-PAGE and visualized with Coomassie brilliant blue staining. M: Molecular weight marker; Lane 1: total bacterial protein; Lane 2: purified protein. (b) PAMPs binding affinity with CgLRRC69. ** means p<0.01, namely, the difference is extremely significant. (c) The binding of LRR domain protein to immobilized LPS as determined by Enzyme-linked immunosorbent assay. Data are expressed as Mean ± SEM (N=3). * means p<0.05, namely, the difference is significant; ** means p<0.01, namely, the difference is extremely significant"
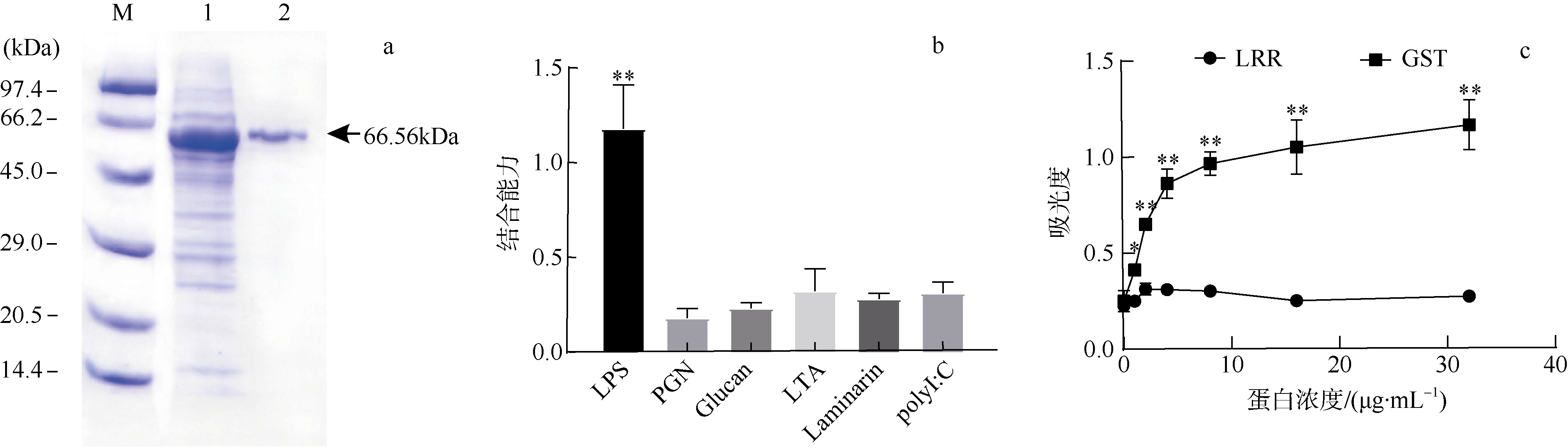

Fig. 6
CgLRRC69 protein with affinity specificity for LPS. (a) The puri?ed CgLRRC69 recombinant protein was analyzed by SDS-PAGE and visualized with Coomassie brilliant blue staining. M: Molecular weight marker; Lane 1: total bacterial protein; Lane 2: purified protein. (b) PAMPs binding affinity with CgLRRC69. ** means p<0.01, namely, the difference is extremely significant. (c) The binding of LRR domain protein to immobilized LPS as determined by Enzyme-linked immunosorbent assay. Data are expressed as Mean ± SEM (N=3). * means p<0.05, namely, the difference is significant; ** means p<0.01, namely, the difference is extremely significant"


Fig. 7
The role of CgLRRC69 in phagocytosis and sterilization. (a) The effects of CgLRRC6 recombinant protein coating on the phagocytic ability of hemocytes. ** means p<0.01, namely, the difference is extremely significant. (b) dsCgLRRC69 knockdown efficiency. Experimental group: dsLRRC69; control group: dsGFP. ** means p<0.01, namely, the difference is extremely significant. (c) Display of the difference in bactericidal ability between the control group and dsCgLRRC69 knockdown treatment group. Data are expressed as Mean ± SEM (N=3). * means p<0.05, namely, the difference is significant"

Fig. 7
The role of CgLRRC69 in phagocytosis and sterilization. (a) The effects of CgLRRC6 recombinant protein coating on the phagocytic ability of hemocytes. ** means p<0.01, namely, the difference is extremely significant. (b) dsCgLRRC69 knockdown efficiency. Experimental group: dsLRRC69; control group: dsGFP. ** means p<0.01, namely, the difference is extremely significant. (c) Display of the difference in bactericidal ability between the control group and dsCgLRRC69 knockdown treatment group. Data are expressed as Mean ± SEM (N=3). * means p<0.05, namely, the difference is significant"

| [1] |
BUCHMANN K, 2014. Evolution of innate immunity: clues from invertebrates via fish to mammals[J]. Frontiers in Immunology, 5:459
doi: 10.3389/fimmu.2014.00459 pmid: 25295041 |
| [2] |
CHAI LIMIN, DAI LINGYUN, CHE YONGZHE, et al, 2009. LRRC19, a novel member of the leucine-rich repeat protein family, activates NF-κB and induces expression of proinflammatory cytokines[J]. Biochemical & Biophysical Research Communications, 388(3):543-548.
pmid: 19679103 |
| [3] | DELLEDONNE M, POLVERARI A, MURGIA I, 2003. The functions of nitric oxide-mediated signaling and changes in gene expression during the hypersensitive response[J]. Antioxidants & Redox Signaling, 5(1):33-41. |
| [4] |
HUANG QINGSONG, YU MINGJIA, CHEN HONGMEI, et al, 2018. LRFN (leucine-rich repeat and fibronectin type-III domain-containing protein) recognizes bacteria and promotes hemocytic phagocytosis in the Pacific oyster Crassostrea gigas[J]. Fish & Shellfish Immunology, 72:622-628.
doi: 10.1016/j.fsi.2017.11.049 pmid: 29190588 |
| [5] |
HUANG SHENGFENG, YUAN SHAOCHUN, GUO LEI, et al, 2008. Genomic analysis of the immune gene repertoire of amphioxus reveals extraordinary innate complexity and diversity[J]. Genome Research, 18(7):1112-1126.
pmid: 18562681 |
| [6] |
JIN M S, LEE J O, 2008. Structures of the toll-like receptor family and its ligand complexes[J]. Immunity, 29(2):182-191.
doi: 10.1016/j.immuni.2008.07.007 pmid: 18701082 |
| [7] |
KAWAI T, AKIRA S, 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors[J]. Nature Immunology, 11(5):373-384.
doi: 10.1038/ni.1863 pmid: 20404851 |
| [8] |
KOBE B, KAJAVA A V, 2001. The leucine-rich repeat as a protein recognition motif[J]. Current Opinion in Structural Biology, 11(6):725-732.
doi: 10.1016/s0959-440x(01)00266-4 pmid: 11751054 |
| [9] |
KUNKEL B N, BROOKS D M, 2002. Cross talk between signaling pathways in pathogen defense[J]. Current Opinion in Plant Biology, 5(4):325-331.
doi: 10.1016/s1369-5266(02)00275-3 pmid: 12179966 |
| [10] | LI YIQUN, SONG XIAORUI, WANG WEILIN, et al, 2017. The hematopoiesis in gill and its role in the immune response of Pacific oyster Crassostrea gigas against secondary challenge with Vibrio splendidus[J]. Developmental & Comparative Immunology, 71:59-69. |
| [11] |
LI ZHAOJIE, ZHANG SHICUI, LIU QINGHUI, 2008. Vitellogenin functions as a multivalent pattern recognition receptor with an opsonic activity[J]. PLoS One, 3(4):e1940.
pmid: 18398466 |
| [12] |
LIU CONGHUI, JIANG SHUAI, WANG MENGQIANG, et al, 2016. A novel siglec (CgSiglec-1) from the Pacific oyster (Crassostrea gigas) with broad recognition spectrum and inhibitory activity to apoptosis, phagocytosis and cytokine release[J]. Developmental & Comparative Immunology, 61:136-144.
pmid: 27032602 |
| [13] |
LIU HOURONG, SONG CHENGWEN, NING JUNHAO, et al, 2020. Identification, functional characterization and the potential role of variable lymphocyte receptor EsVLRA from Eriocheir sinensis in response to secondary challenge after Vibrio parahaemolyticus vaccine[J]. Fish & Shellfish Immunology, 98:201-209.
doi: 10.1016/j.fsi.2020.01.011 pmid: 31923564 |
| [14] |
MATSUSHIMA N, TAKATSUKA S, MIYASHITA H, et al, 2019. Leucine rich repeat proteins: sequences, mutations, structures and diseases[J]. Protein & Peptide Letters, 26(2):108-131.
doi: 10.2174/0929866526666181208170027 pmid: 30526451 |
| [15] |
MEDZHITOV R, 2007. Recognition of microorganisms and activation of the immune response[J]. Nature, 449(7164):819-826.
pmid: 17943118 |
|
MEDZHITOV R, 2007. Recognition of microorganisms and activation of the immune response[J]. Nature, 449(7164):819-826.
pmid: 17943118 |
|
| [16] |
NG A, XAVIER R J, 2011a. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity[J]. Autophagy, 7(9):1082-1084.
pmid: 21606681 |
|
NG A, XAVIER R J, 2011a. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity[J]. Autophagy, 7(9):1082-1084.
pmid: 21606681 |
|
| [17] | NG A C Y, EISENBERG J M, HEATH R J W, et al, 2011b. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity[J]. Proceedings of the National Academy of Sciences of the United States of America, 108(S1):4631-4638. |
| NG A C Y, EISENBERG J M, HEATH R J W, et al, 2011b. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity[J]. Proceedings of the National Academy of Sciences of the United States of America, 108(S1):4631-4638. | |
| [18] |
PÅLSSON-MCDERMOTT E M, O'NEILL L A J, 2007. Building an immune system from nine domains[J]. Biochemical Society Transactions, 35(Pt 6):1437-1444.
pmid: 18031241 |
|
PÅLSSON-MCDERMOTT E M, O'NEILL L A J, 2007. Building an immune system from nine domains[J]. Biochemical Society Transactions, 35(Pt 6):1437-1444.
pmid: 18031241 |
|
| [19] |
PEART J R, MESTRE P, LU RUI, et al, 2005. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus[J]. Current Biology, 15(10):968-973.
pmid: 15916955 |
|
PEART J R, MESTRE P, LU RUI, et al, 2005. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus[J]. Current Biology, 15(10):968-973.
pmid: 15916955 |
|
| [20] |
ROTHBERG J M, JACOBS J R, GOODMAN C S, et al, 1990. slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains[J]. Genes & Development, 4:2169-2187.
doi: 10.1101/gad.4.12a.2169 pmid: 2176636 |
|
ROTHBERG J M, JACOBS J R, GOODMAN C S, et al, 1990. slit: an extracellular protein necessary for development of midline glia and commissural axon pathways contains both EGF and LRR domains[J]. Genes & Development, 4:2169-2187.
doi: 10.1101/gad.4.12a.2169 pmid: 2176636 |
|
| [21] | SHOKAL U, ELEFTHERIANOS I, 2017. Evolution and function of thioester-containing proteins and the complement system in the innate immune response[J]. Frontiers in Immunology, 8(1):759. |
| SHOKAL U, ELEFTHERIANOS I, 2017. Evolution and function of thioester-containing proteins and the complement system in the innate immune response[J]. Frontiers in Immunology, 8(1):759. | |
| [22] |
SITARAM N, 2006. Antimicrobial peptides with unusual amino acid compositions and unusual structures[J]. Current Medicinal Chemistry, 13(6):679-696.
doi: 10.2174/092986706776055689 pmid: 16529559 |
|
SITARAM N, 2006. Antimicrobial peptides with unusual amino acid compositions and unusual structures[J]. Current Medicinal Chemistry, 13(6):679-696.
doi: 10.2174/092986706776055689 pmid: 16529559 |
|
| [23] |
SRIPHAIJIT T, SENAPIN S, 2007. High expression of a novel leucine-rich repeat protein in hemocytes and the lymphoid organ of the black tiger shrimp Penaeus monodon[J]. Fish & Shellfish Immunology, 22(3):264-271.
pmid: 16926101 |
|
SRIPHAIJIT T, SENAPIN S, 2007. High expression of a novel leucine-rich repeat protein in hemocytes and the lymphoid organ of the black tiger shrimp Penaeus monodon[J]. Fish & Shellfish Immunology, 22(3):264-271.
pmid: 16926101 |
|
| [24] |
VAN LOOKEREN CAMPAGNE M, WIESMANN C, BROWN E J, 2007. Macrophage complement receptors and pathogen clearance[J]. Cellular Microbiology, 9(9):2095-2102.
doi: 10.1111/j.1462-5822.2007.00981.x pmid: 17590164 |
|
VAN LOOKEREN CAMPAGNE M, WIESMANN C, BROWN E J, 2007. Macrophage complement receptors and pathogen clearance[J]. Cellular Microbiology, 9(9):2095-2102.
doi: 10.1111/j.1462-5822.2007.00981.x pmid: 17590164 |
|
| [25] |
WANG MENGQIANG, WANG LINGLING, GUO YING, et al, 2016a. An LRR-only protein representing a new type of pattern recognition receptor in Chlamys farreri[J]. Developmental & Comparative Immunology, 54(1):145-155.
doi: 10.1016/j.dci.2015.09.006 pmid: 26385592 |
|
WANG MENGQIANG, WANG LINGLING, GUO YING, et al, 2016a. An LRR-only protein representing a new type of pattern recognition receptor in Chlamys farreri[J]. Developmental & Comparative Immunology, 54(1):145-155.
doi: 10.1016/j.dci.2015.09.006 pmid: 26385592 |
|
| [26] |
WANG MENGQIANG, WANG LINGLING, XIN LUSHENG, et al, 2016b. Two novel LRR-only proteins in Chlamys farreri: Similar in structure, yet different in expression profile and pattern recognition[J]. Developmental & Comparative Immunology, 59:99-109.
doi: 10.1016/j.dci.2016.01.013 pmid: 26826425 |
|
WANG MENGQIANG, WANG LINGLING, XIN LUSHENG, et al, 2016b. Two novel LRR-only proteins in Chlamys farreri: Similar in structure, yet different in expression profile and pattern recognition[J]. Developmental & Comparative Immunology, 59:99-109.
doi: 10.1016/j.dci.2016.01.013 pmid: 26826425 |
|
| [27] |
WANG XIALU, ZHANG YUEQI, ZHANG RONG, et al, 2019. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens[J]. Current Opinion in Insect Science, 33:105-110.
doi: 10.1016/j.cois.2019.05.004 pmid: 31358188 |
|
WANG XIALU, ZHANG YUEQI, ZHANG RONG, et al, 2019. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens[J]. Current Opinion in Insect Science, 33:105-110.
doi: 10.1016/j.cois.2019.05.004 pmid: 31358188 |
|
| [28] | WANG XIANWEI, GAO JIE, XU YIHUI, et al, 2017b. Novel pattern recognition receptor protects shrimp by preventing bacterial colonization and promoting phagocytosis[J]. Journal of Immunology, 198(8):3045-3057. |
| WANG XIANWEI, GAO JIE, XU YIHUI, et al, 2017b. Novel pattern recognition receptor protects shrimp by preventing bacterial colonization and promoting phagocytosis[J]. Journal of Immunology, 198(8):3045-3057. | |
| [29] |
WANG XIUDAN, WANG MENGQIANG, XU QINGSONG, et al, 2017c. Two novel LRR and Ig domain-containing proteins from oyster Crassostrea gigas function as pattern recognition receptors and induce expression of cytokines[J]. Fish & Shellfish Immunology, 70:308-318.
doi: 10.1016/j.fsi.2017.09.023 pmid: 28889011 |
|
WANG XIUDAN, WANG MENGQIANG, XU QINGSONG, et al, 2017c. Two novel LRR and Ig domain-containing proteins from oyster Crassostrea gigas function as pattern recognition receptors and induce expression of cytokines[J]. Fish & Shellfish Immunology, 70:308-318.
doi: 10.1016/j.fsi.2017.09.023 pmid: 28889011 |
|
| [30] |
ZHANG LINLIN, LI LI, ZHANG GUOFAN, 2011. A Crassostrea gigas Toll-like receptor and comparative analysis of TLR pathway in invertebrates[J]. Fish & Shellfish Immunology, 30(2):653-660.
doi: 10.1016/j.fsi.2010.12.023 pmid: 21195773 |
|
ZHANG LINLIN, LI LI, ZHANG GUOFAN, 2011. A Crassostrea gigas Toll-like receptor and comparative analysis of TLR pathway in invertebrates[J]. Fish & Shellfish Immunology, 30(2):653-660.
doi: 10.1016/j.fsi.2010.12.023 pmid: 21195773 |
|
| [31] |
ZHANG YANG, HE XIAOCUI, YU FENG, et al, 2013. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity[J]. PLoS One, 8(10):e76464.
doi: 10.1371/journal.pone.0076464 pmid: 24098508 |
|
ZHANG YANG, HE XIAOCUI, YU FENG, et al, 2013. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity[J]. PLoS One, 8(10):e76464.
doi: 10.1371/journal.pone.0076464 pmid: 24098508 |
| [1] | YANG Liuqinqing, CHU Moxian, LIU Bilin, KONG Xianghong. Pattern recognition of Sthenoteuthis oualaniensis based on BPNN about momentum and self-adaption [J]. Journal of Tropical Oceanography, 2021, 40(6): 102-110. |
|
||