Journal of Tropical Oceanography ›› 2024, Vol. 43 ›› Issue (6): 145-159.doi: 10.11978/2023196CSTR: 32234.14.2023196
• Marine Biology • Previous Articles Next Articles
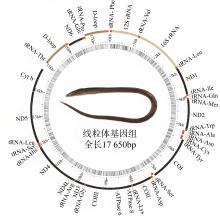
The mitochondrial genomic characteristics and phylogenetic relationship analysis of Pisodonophis cancrivorus (Anguilliformes, Ophichthidae)
YANG Lan( ), ZHAO Yao, LIU Yuping, YANG Tianyan(
), ZHAO Yao, LIU Yuping, YANG Tianyan( )
)
- School of Fisheries, Zhejiang Ocean University, Zhoushan 316022, China
-
Received:2023-12-15Revised:2024-02-11Online:2024-11-10Published:2024-12-05 -
Contact:YANG Tianyan -
Supported by:Province Key Research and Development Program of Zhejiang(2021C2047); Technology Planning Project of Zhoushan(2022C41022); National Innovation and Entrepreneurship Training Program for College Students(202310340056); Science and Technology Innovation Project of College Students in Zhejiang Province(2023R411005); Innovation Project of College Students in Zhejiang Ocean University(2023-A-025)
CLC Number:
- S917.4
Cite this article
YANG Lan, ZHAO Yao, LIU Yuping, YANG Tianyan. The mitochondrial genomic characteristics and phylogenetic relationship analysis of Pisodonophis cancrivorus (Anguilliformes, Ophichthidae)[J].Journal of Tropical Oceanography, 2024, 43(6): 145-159.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
The mitogenome annotation of P. cancrivorus"
| 基因 | 编码链 | 起始位点 | 终止位点 | 长度/bp | 基因间隔/bp | 编码氨基酸数量 | 起始密码子 | 终止密码子 | 反密码子 |
|---|---|---|---|---|---|---|---|---|---|
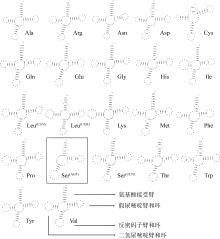
| tRNA-Phe(F) | H | 1 | 68 | 68 | 0 | GAA | |||
| 12S rRNA | H | 69 | 1 027 | 959 | 0 | ||||
| tRNA-Val(V) | H | 1 028 | 1 098 | 71 | 0 | TAC | |||
| 16S rRNA | H | 1 099 | 2 799 | 1 701 | 0 | ||||
| tRNA-LeuUUR(L) | H | 2 800 | 2 875 | 76 | 0 | TAA | |||
| ND1 | H | 2 876 | 3 844 | 969 | 0 | 322 | ATG | TAA | |
| tRNA-Ile(I) | H | 3 847 | 3 919 | 73 | 2 | GAT | |||
| tRNA-Gln(Q) | L | 3 919 | 3 989 | 71 | -1 | TTG | |||
| tRNA-Met(M) | H | 3 989 | 4 057 | 69 | -1 | CAT | |||
| ND2 | H | 4 058 | 5 114 | 1 057 | 0 | 352 | ATG | T-- | |
| tRNA-Trp(W) | H | 5 115 | 5 183 | 69 | 0 | TCA | |||
| tRNA-Ala(A) | L | 5 185 | 5 253 | 69 | 1 | TGC | |||
| tRNA-Asn(N) | L | 5 255 | 5 327 | 73 | 1 | GTT | |||
| OL | H | 5 329 | 5 349 | 21 | 1 | ||||
| tRNA-Cys(C) | L | 5 371 | 5 437 | 67 | 21 | GCA | |||
| tRNA-Tyr(Y) | L | 5 438 | 5 509 | 72 | 0 | GTA | |||
| COI | H | 5 511 | 7 151 | 1 641 | 1 | 546 | GTG | TAA | |
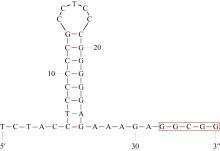
| tRNA-SerUCN(S) | L | 7 169 | 7 239 | 71 | 17 | TGA | |||
| tRNA-Asp(D) | H | 7 245 | 7 312 | 68 | 5 | GTC | |||
| COII | H | 7 319 | 8 009 | 691 | 6 | 230 | ATG | T-- | |
| tRNA-Lys(K) | H | 8 010 | 8 084 | 75 | 0 | TTT | |||
| ATP8 | H | 8 086 | 8 253 | 168 | 1 | 55 | ATG | TAA | |
| ATP6 | H | 8 244 | 8 923 | 680 | -10 | 226 | ATG | TA- | |
| COIII | H | 8 924 | 9 708 | 785 | 0 | 261 | ATG | TA- | |
| tRNA-Gly(G) | H | 9 709 | 9 780 | 72 | 0 | TCC | |||
| ND3 | H | 9 781 | 10 129 | 349 | 0 | 116 | ATG | T-- | |
| tRNA-Arg(R) | H | 10 130 | 10 199 | 70 | 0 | TCG | |||
| ND4L | H | 10 200 | 10 496 | 297 | 0 | 98 | ATG | TAA | |
| ND4 | H | 10 490 | 11 870 | 1 381 | -7 | 610 | ATG | T-- | |
| tRNA-His(H) | H | 11 871 | 11 939 | 69 | 0 | GTG | |||
| tRNA-SerAGY(S) | H | 11 940 | 12 009 | 70 | 0 | GCT | |||
| tRNA-LeuCUN(L) | H | 12 010 | 12 082 | 73 | 0 | TAG | |||
| ND5 | H | 12 083 | 13 918 | 1 836 | 0 | 611 | ATG | TAG | |
| Cyt b | H | 13 933 | 15 073 | 1 141 | 14 | 380 | ATG | T-- | |
| tRNA-Thr(T) | H | 15 074 | 15 145 | 72 | 0 | TGT | |||
| D-loop1 | H | 15 146 | 16 111 | 966 | 0 | ||||
| ND6 | L | 16 112 | 16 630 | 519 | 0 | 172 | ATG | TAG | |
| tRNA-Glu(E) | L | 16 631 | 16 699 | 69 | 0 | TTC | |||
| tRNA-Pro(P) | L | 16 703 | 16 775 | 73 | 3 | TGG | |||
| D-loop2 | H | 16 776 | 17 650 | 875 | 0 |
Tab. 2
The base compositions and skewness in the mitogenome of P. cancrivorus"
| 基因/区域 | 长度/bp | A/% | T/% | G/% | C/% | A+T/% | AT偏倚 | GC偏倚 |
|---|---|---|---|---|---|---|---|---|
| 线粒体全序列 | 17 650 | 31.72 | 26.26 | 15.58 | 26.44 | 57.98 | 0.094 | -0.259 |
| 13个蛋白质编码基因 | 11 514 | 29.33 | 28.23 | 15.46 | 26.98 | 57.56 | 0.019 | -0.271 |
| 密码子第1位点 | 3 849 | 28.55 | 23.33 | 22.79 | 25.33 | 51.88 | 0.101 | -0.053 |
| 密码子第2位点 | 3 831 | 23.52 | 35.47 | 13.94 | 27.07 | 58.99 | -0.203 | -0.320 |
| 密码子第3位点 | 3 834 | 35.92 | 25.93 | 9.62 | 28.53 | 61.85 | 0.162 | -0.496 |
| ND1 | 969 | 27.86 | 27.86 | 15.38 | 28.90 | 55.72 | 0.000 | -0.305 |
| ND2 | 1 057 | 33.59 | 24.50 | 13.62 | 28.29 | 58.09 | 0.156 | -0.350 |
| COI | 1 641 | 28.28 | 29.25 | 17.55 | 24.92 | 57.53 | -0.017 | -0.174 |
| COII | 691 | 30.82 | 26.19 | 15.63 | 27.36 | 57.01 | 0.081 | -0.273 |
| ATP8 | 168 | 33.33 | 25.00 | 10.12 | 31.55 | 58.33 | 0.143 | -0.514 |
| ATP6 | 680 | 29.12 | 28.97 | 12.35 | 29.56 | 58.09 | 0.003 | -0.411 |
| COIII | 785 | 28.15 | 27.90 | 17.20 | 26.75 | 56.05 | 0.004 | -0.217 |
| ND3 | 349 | 28.37 | 31.52 | 14.04 | 26.07 | 59.89 | -0.053 | -0.300 |
| ND4L | 297 | 28.28 | 26.60 | 13.47 | 31.65 | 54.88 | 0.031 | -0.403 |
| ND4 | 1 381 | 29.98 | 27.59 | 13.76 | 28.67 | 57.57 | 0.042 | -0.351 |
| ND5 | 1 836 | 32.57 | 26.85 | 12.91 | 27.67 | 59.42 | 0.096 | -0.364 |
| Cyt b | 1 141 | 28.66 | 29.71 | 15.07 | 26.56 | 58.37 | -0.018 | -0.276 |
| ND6 | 519 | 38.73 | 15.03 | 14.06 | 32.18 | 53.76 | 0.441 | -0.392 |
| 12S rRNA | 959 | 33.58 | 21.38 | 20.02 | 25.02 | 54.96 | 0.222 | -0.111 |
| 16S rRNA | 1 701 | 37.92 | 19.05 | 18.99 | 24.04 | 56.97 | 0.331 | -0.117 |
| D-loop1 | 966 | 32.19 | 32.51 | 14.29 | 21.01 | 64.70 | 0.441 | -0.392 |
| D-loop2 | 875 | 36.23 | 29.60 | 10.74 | 23.43 | 65.83 | 0.441 | -0.392 |
Tab. 3
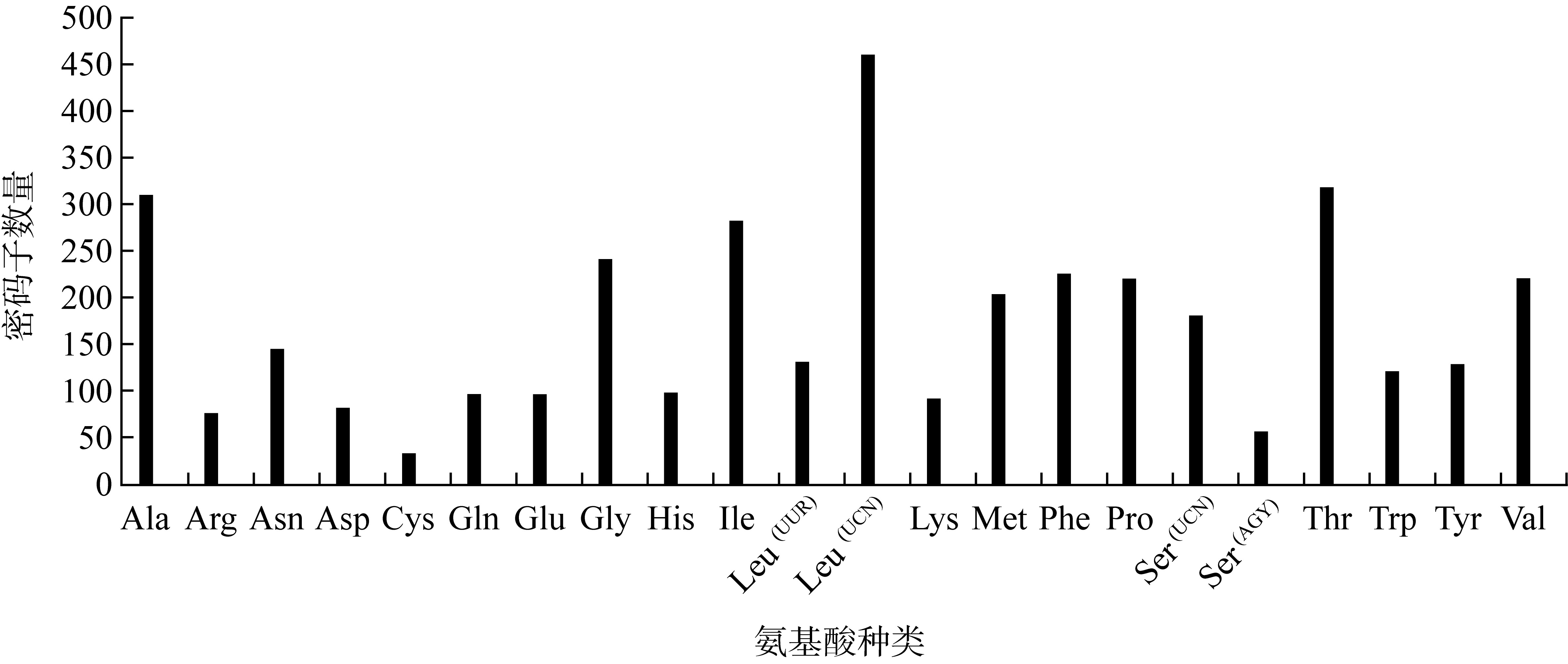
Frequency of codon usages in 13 PCGs for P. cancrivorus"
| 密码子 | 计数 | RSCU | 密码子 | 计数 | RSCU | 密码子 | 计数 | RSCU | 密码子 | 计数 | RSCU |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UUU(F) | 121 | 1.07 | UCU(S) | 30 | 0.76 | UAU(Y) | 62 | 0.96 | UGU(C) | 10 | 0.61 |
| UUC(F) | 105 | 0.93 | UCC(S) | 63 | 1.59 | UAC(Y) | 67 | 1.04 | UGC(C) | 23 | 1.39 |
| UUA(L) | 101 | 1.02 | UCA(S) | 84 | 2.12 | UAA(*) | 0 | 0.00 | UGA(W) | 108 | 1.79 |
| UUG(L) | 31 | 0.31 | UCG(S) | 4 | 0.10 | UAG(*) | 1 | 0.80 | UGG(W) | 13 | 0.21 |
| CUU(L) | 108 | 1.09 | CCU(P) | 32 | 0.58 | CAU(H) | 33 | 0.67 | CGU(R) | 11 | 0.58 |
| CUC(L) | 108 | 1.09 | CCC(P) | 67 | 1.22 | CAC(H) | 66 | 1.33 | CGC(R) | 10 | 0.53 |
| CUA(L) | 190 | 1.92 | CCA(P) | 107 | 1.95 | CAA(Q) | 84 | 1.75 | CGA(R) | 46 | 2.42 |
| CUG(L) | 55 | 0.56 | CCG(P) | 14 | 0.25 | CAG(Q) | 12 | 0.25 | CGG(R) | 9 | 0.47 |
| AUU(I) | 202 | 1.43 | ACU(T) | 48 | 0.6 | AAU(N) | 53 | 0.73 | AGU(S) | 13 | 0.33 |
| AUC(I) | 81 | 0.57 | ACC(T) | 100 | 1.25 | AAC(N) | 92 | 1.27 | AGC(S) | 44 | 1.11 |
| AUA(M) | 155 | 1.52 | ACA(T) | 159 | 1.99 | AAA(K) | 79 | 1.72 | AGA(*) | 2 | 1.60 |
| AUG(M) | 49 | 0.48 | ACG(T) | 12 | 0.15 | AAG(K) | 13 | 0.28 | AGG(*) | 2 | 1.60 |
| GUU(V) | 59 | 1.07 | GCU(A) | 53 | 0.68 | GAU(D) | 25 | 0.61 | GGU(G) | 28 | 0.46 |
| GUC(V) | 48 | 0.87 | GCC(A) | 129 | 1.66 | GAC(D) | 57 | 1.39 | GGC(G) | 64 | 1.06 |
| GUA(V) | 89 | 1.61 | GCA(A) | 120 | 1.55 | GAA(E) | 79 | 1.65 | GGA(G) | 118 | 1.96 |
| GUG(V) | 25 | 0.45 | GCG(A) | 8 | 0.10 | GAG(E) | 17 | 0.35 | GGG(G) | 31 | 0.51 |
| [1] |
陈慧林, 朱永航, 范海页, 等, 2023. 基于18S和28S rDNA基因的房舍甲螨DNA条形码比较研究[J]. 中国寄生虫学与寄生虫病杂志, 41(2): 163-169.
doi: 10.12140/j.issn.1000-7423.2023.02.006 |
|
doi: 10.12140/j.issn.1000-7423.2023.02.006 |
|
| [2] |
陈治, 高天翔, 2023. 线粒体12S与COI条形码对海洋鱼类的鉴定差异[J]. 海南热带海洋学院学报, 30(2): 10-16.
|
|
|
|
| [3] |
邓家刚, 2008. 广西海洋药物[M]. 南宁: 广西科学技术出版社.
|
|
|
|
| [4] |
董江星, 时伟, 孔晓瑜, 等, 2018. 褐斜鲽(Plagiopsetta glossa)线粒体基因组特征及重排机制研究[J]. 热带海洋学报, 37(1): 1-11.
doi: 10.11978/2017041 |
|
|
|
| [5] |
龚理, 时伟, 司李真, 等, 2013. 鱼类线粒体DNA重排研究进展[J]. 动物学研究, 34(6): 666-673.
|
|
|
|
| [6] |
郭新红, 刘少军, 刘巧, 等, 2004. 鱼类线粒体DNA研究新进展[J]. 遗传学报, 31(9): 983-1000.
|
|
|
|
| [7] |
连总强, 滚双宝, 李力, 等, 2017. 基于第二代测序技术兰州鲇线粒体基因组全序列测定与分析[J]. 水生生物学报, 41(2): 334-345.
|
|
|
|
| [8] |
梁日深, 杨杰銮, 谢瑞琳, 等, 2022. 巨石斑鱼与斜带石斑鱼线粒体基因组测序及物种有效性分析[J]. 中国海洋大学学报, 52(6): 50-61.
|
|
|
|
| [9] |
林龙山, 张静, 宋普庆, 等, 2013. 东山湾及其邻近海域常见游泳动物[M]. 北京: 海洋出版社.
|
|
|
|
| [10] |
刘东, 2005. 中国蛇鳗科鱼类嗅觉器官的比较形态学及其系统发育研究[D]. 上海: 上海海洋大学.
|
|
|
|
| [11] |
刘焕章, 2004. 用mtDNA 12S rRNA序列变异检验鲤形目鱼类系统发育关系[J]. 遗传学报, 31(2): 137-142.
|
|
|
|
| [12] |
刘继兵, 赵洪喜, 2022. 基于18S rRNA的胎毛滴虫感染PCR诊断方法的建立[J]. 中国寄生虫学与寄生虫病杂志, 40(5): 682-685.
doi: 10.12140/j.issn.1000-7423.2022.05.019 |
|
|
|
| [13] |
刘凯, 冯晓宇, 马恒甲, 等, 2020. 钱塘江三角鲂线粒体基因组测序及其结构特征分析[J]. 浙江农业学报, 32(9): 1591-1608.
doi: 10.3969/j.issn.1004-1524.2020.09.08 |
|
doi: 10.3969/j.issn.1004-1524.2020.09.08 |
|
| [14] |
刘文强, 贾玉萍, 赵宏坤, 2006. 16 S rRNA在细菌分类鉴定研究中的应用[J]. 动物医学进展, 27(11): 15-18.
|
|
|
|
| [15] |
刘毅敏, 赵先英, 王祥智, 等, 2006. 基于自由能的生物大分子结构研究[J]. 化学世界, 47(8): 508-510.
|
|
|
|
| [16] |
宁子君, 刘玉萍, 张书飞, 等, 2022. 艾氏蛇鳗线粒体基因组全序列结构分析和系统发育关系探讨[J]. 中国水产科学, 29(9): 1264-1276.
|
|
|
|
| [17] |
钱立富, 2018. 蛇类线粒体基因组结构演化研究及原矛头蝮属生物地理学分析[D]. 合肥: 安徽大学.
|
|
|
|
| [18] |
申欣, 田美, 孟学平, 等, 2014. 鳗鲡目鱼类线粒体蛋白质编码基因易位及系统演化关系分析[J]. 海洋学报, 36(4): 73-81.
|
|
|
|
| [19] |
唐文乔, 张春光, 2004. 蛇鳗科分类综述及中国蛇鳗科系统分类(鱼纲, 鳗鲡目)[J]. 上海水产大学学报, 13(1): 16-22.
|
|
|
|
| [20] |
唐优良, 章群, 余帆洋, 等, 2011. 基于12S rRNA部分序列分析的中国8种笛鲷科鱼类系统发育初探[J]. 海洋科学, 35(2): 22-26.
|
|
|
|
| [21] |
杨婧, 黄原, 2016. 线粒体基因组的高通量测序策略[J]. 生命科学, 28(1): 112-117.
|
|
|
|
| [22] |
张燕萍, 郑红梅, 邵芳, 等, 2016. 沙塘鳢线粒体基因组全序列的测定和分析[J]. 常熟理工学院学报(自然科学), 30(4): 97-104.
|
|
|
|
| [23] |
张稚兰, 林汝榕, 邢炳鹏, 2017. COI基因序列在蛇鳗科鱼类种类鉴定中的适用性研究[J]. 应用海洋学学报, 36(3): 411-416.
|
|
|
|
| [24] |
钟东, 赵贵军, 张振书, 等, 2002. 基因组内碱基分布整体均衡与局部不均衡的研究进展[J]. 遗传, 24(3): 351-355.
|
|
|
|
| [25] |
周慧琦, 2014. 基因组GC含量与碱基、密码子和氨基酸使用偏好的关系[D]. 成都: 电子科技大学.
|
|
|
|
| [26] |
邹新慧, 葛颂, 2008. 基因树冲突与系统发育基因组学研究[J]. 植物分类学报, 46(6): 795-807.
|
|
|
|
| [27] |
|
| [28] |
pmid: 6284948 |
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
doi: 10.1093/nar/gkh340 pmid: 15034147 |
| [34] |
doi: 10.1111/j.1558-5646.1985.tb00420.x pmid: 28561359 |
| [35] |
|
| [36] |
|
| [37] |
doi: 10.1080/10635150390235520 pmid: 14530136 |
| [38] |
doi: 10.1098/rspb.2002.2218 pmid: 12614582 |
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
doi: 10.1093/bioinformatics/17.8.754 pmid: 11524383 |
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
pmid: 12949142 |
| [47] |
doi: 10.1093/molbev/mst141 pmid: 23955518 |
| [48] |
pmid: 7508516 |
| [49] |
|
| [50] |
pmid: 7608989 |
| [51] |
|
| [52] |
|
| [53] |
doi: 10.1093/nar/25.5.955 pmid: 9023104 |
| [54] |
|
| [55] |
doi: 10.1186/1471-2164-12-84 pmid: 21276253 |
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
pmid: 12470944 |
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
doi: 10.1007/BF00186547 pmid: 7563121 |
| [66] |
doi: 10.1093/molbev/msn083 pmid: 18397919 |
| [67] |
|
| [68] |
|
| [69] |
pmid: 8015429 |
| [70] |
|
| [71] |
doi: 10.1186/1471-2148-13-173 pmid: 23962312 |
| [72] |
|
| [73] |
doi: 10.1093/molbev/msab120 pmid: 33892491 |
| [74] |
|
| [75] |
|
| [76] |
doi: 10.1093/oxfordjournals.molbev.a025661 pmid: 8752002 |
| [77] |
|
| [1] | HUO Jiaxin, LI Yingxin, SONG Yan, ZHU Qing, ZHOU Weihua, YUAN Xiangcheng, HUANG Hui, LIU Sheng. Complete mitochondrial genome of Cladopsammia gracilis and Rhizopsammia wettsteini (Scleractinia, Dendrophylliidae) and its phylogenetic implications* [J]. Journal of Tropical Oceanography, 2024, 43(3): 22-30. |
| [2] | HUANG Peixian, YAO Xuemei, YU Qiaochi, ZHANG Jiayu. Mitogenome characteristics and phylogenetic analysis of Siphonosoma australe in Hainan [J]. Journal of Tropical Oceanography, 2023, 42(2): 54-63. |
| [3] | LONG Chao, LUO Zhaohe, WEI Zhangliang, YANG Fangfang, LI Ru, LONG Lijuan. Morphology and phylogeny of zooxanthellae Effrenium voratum from Luhuitou reef in Sanya, Hainan province [J]. Journal of Tropical Oceanography, 2021, 40(4): 35-43. |
| [4] | NIU Biaobiao, ZHAI Mengyi, LI Yang. Morphological and Phylogenetic Studies on Chaetoceros seychellarus [J]. Journal of Tropical Oceanography, 2021, 40(4): 44-49. |
| [5] | Lifen HUANG, Qun LI, Songhui LÜ, Liang ZHANG, Xuedong XIE. Morphology, phylogeny and toxicity of Coolia tropicalis (Dinophyceae) from the Xisha Islands, China [J]. Journal of Tropical Oceanography, 2020, 39(3): 86-97. |
| [6] | Lichun JIANG, Qun LI, Songhui LÜ. Morphology and Phylogenetics study on the species of Amphidinium (Gymnodinials, Dinophyceae) from Weizhou Island, Guangxi [J]. Journal of Tropical Oceanography, 2020, 39(3): 106-115. |
| [7] | Bin CHEN, Wei HE, Haitao LI, Peng WU, Yuzhang Xiao, Xiangli LYU, Jing HE. Identification and phylogenetic analysis of Sagittidae (Chaetognatha) in the South China Sea based on mitochondrial CO#cod#x02160; gene [J]. Journal of Tropical Oceanography, 2017, 36(6): 100-108. |
| [8] | FAN Cheng-hui, LIU Li, SHEN Cheng, GUO Yu-song. Molecular cloning and sequence analysis of the copper-zinc superoxide dismutase gene in Galaxea astreata [J]. Journal of Tropical Oceanography, 2015, 34(1): 83-89. |
| [9] | LONG Chao, CHEN Xian-yun, CHEN Bo, HE Bi-juan, GAO Cheng-hai, WANG Yi-bing. Morphological and phylogenetic analysis of Prorocentrum triestinum isolated from the Beibu Gulf [J]. Journal of Tropical Oceanography, 2014, 33(2): 66-71. |
| [10] | HU Yong-le,LIANG Xu-fang,WANG Lin,LI Guan-gui,LIU Xiu-xia,WANG Yun-xin. Molecular cloning and sequence analysis of pancreatic trypsinogen and amylase from orange-spotted grouper (Epinephelus coioides) [J]. Journal of Tropical Oceanography, 2010, 29(5): 125-131. |
| [11] | LIANG Ri-shen,YANG Guo-hua,LUO Da-ji,ZHONG Shan,ZOU Ji-xing. Phylogenetic relationship of 10 species in haemulidae based on S7 ribosomal protein gene [J]. Journal of Tropical Oceanography, 2010, 29(5): 98-102. |
| [12] | MA Ling-bo,MA Chun-yan,ZHANG Feng-ying,JIANG Ke-ji. Phylogenetic relationships of four Scylla species based on three kinds of mitochondrial sequences [J]. Journal of Tropical Oceanography, 2010, 29(5): 88-93. |
|
||