Journal of Tropical Oceanography ›› 2022, Vol. 41 ›› Issue (6): 44-55.doi: 10.11978/2022061CSTR: 32234.14.2022061
• Special Column: Mangrove Forest • Previous Articles Next Articles
Expression of DEAD-box RNA helicase enzyme genes in Lumnitzera littorea under low temperature stress
HAO Lulu1,2( ), KE Mingsi2, ZHU Yixiu2, XU Yanmin2, ZHANG Ying2,3, ZHENG Chunfang1(
), KE Mingsi2, ZHU Yixiu2, XU Yanmin2, ZHANG Ying2,3, ZHENG Chunfang1( )
)
- 1. College of Life and Environmental Science, Wenzhou University, Wenzhou 325035, China
2. School of Life Sciences and Technology, Lingnan Normal University, Zhanjiang 524048, China
3. Forestry Science Research of Hainan Province (Hainan Mangrove Research Institute), Haikou 571129, China
-
Received:2022-03-29Revised:2022-05-23Online:2022-11-10Published:2022-05-31 -
Contact:ZHENG Chunfang E-mail:1973497638@qq.com;20195101@wzu.edu.cn -
Supported by:Basic scientific research work of Hainan Forestry Research Institute (Hainan Mangrove Research Institute)(KYYS-2021-04);Technological Innovation Special project of Hainan Scientific Research Institute(KYYS-2021-13);Technological Innovation Special project of Hainan Scientific Research Institute(KYYS-2021-22);Special Basic Research Work Project for Technological Innovation in Hainan Research Institutes(jcxk202003);National Natural Science Foundation of China(32071503)
CLC Number:
- Q943.2
Cite this article
HAO Lulu, KE Mingsi, ZHU Yixiu, XU Yanmin, ZHANG Ying, ZHENG Chunfang. Expression of DEAD-box RNA helicase enzyme genes in Lumnitzera littorea under low temperature stress[J].Journal of Tropical Oceanography, 2022, 41(6): 44-55.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab.1
Functional website for online analysis and website link"
| 网站 | 功能 | 网址链接 | 参数设置 |
|---|---|---|---|
| ExPASy ProtParam | 理化性质分析 | https://web.expasy.org/protparam/ | 默认参数 |
| PSORT | 亚细胞定位分析 | https://www.genscript.com/psort.html?src=leftbar | 默认参数 |
| ProtScale | 亲水性分析 | https://web.expasy.org/cgi-bin/protscale/protscale.pl | Hphob./Kyte & Doolittle, Window size: 9; Weight variation model:linear |
| SOPMA | 二级结构分析 | https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html | Number of conformational states: 4; Similarity threshold: 8; Window width: 17 |
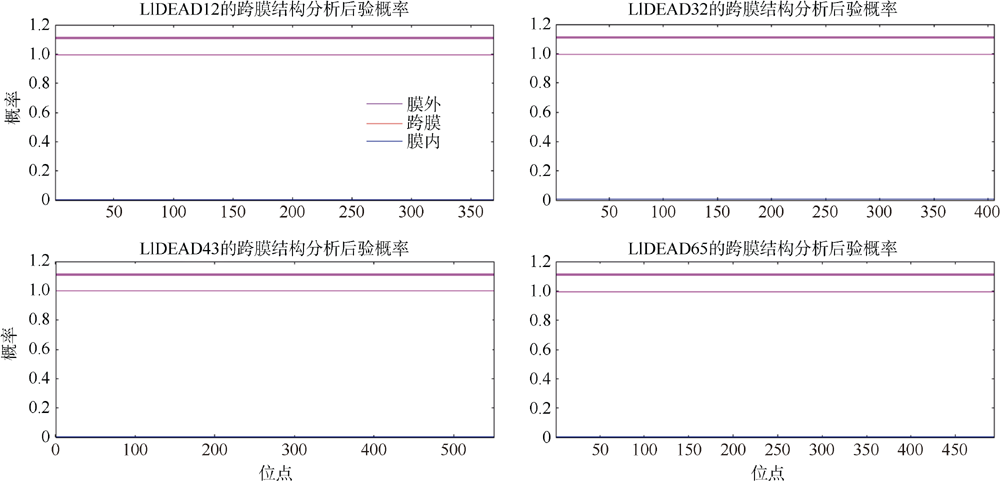
| TMHMM | 跨膜结构分析 | https://services.healthtech.dtu.dk/service.php?TMHMM-2.0 | 默认参数 |
| SWISS-MODEL | 三级结构分析 | https://swissmodel.expasy.org/interactive | 默认参数 |
| SignalP4.1 | 信号肽分析 | https://services.healthtech.dtu.dk/service.php?SignalP-4.1 | Organism group: Eukaryotes; D-cutoff values: Default; Output format: Standard; Method: Input sequences may include TM regions |
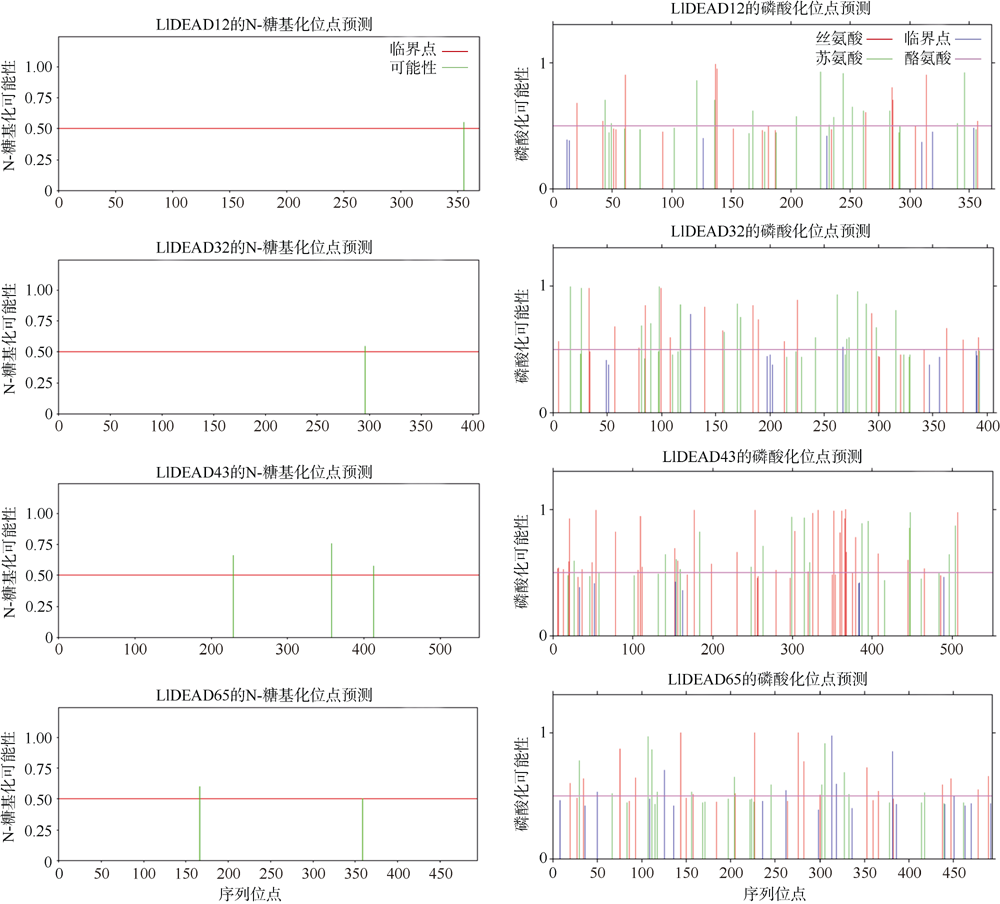
| NetPhos3.1 | 磷酸化位点分析 | https://services.healthtech.dtu.dk/service.php?NetPhos-3.1 | Residues to predict: all three; |
| NetNGlyc1.0 | 糖基化位点分析 | https://services.healthtech.dtu.dk/service.php?NetNGlyc-1.0 | 默认设置 |
Tab. 2
Primer sequence of qPCR"
| 引物 | 引物序列(5′-3′) | 引物长度/bp |
|---|---|---|
| AthU6-F | ACATCCGATAAAATTGGAACGA | 168 |
| AthU6-R | TTTTTTTGGACCATTTCTCGAT | |
| LlDEAD12-F | GGCTATGACTACCGCAATCTC | 237 |
| LlDEAD12-R | GCCCTTTCAACACTATGCTTCT | |
| LlDEAD32-F | CTGTTGTGCTCTGCCCCA | 204 |
| LlDEAD32-R | CCATGTTCCCATCTTCAATATG | |
| LlDEAD43-F | ATCCTTCCATAGGCGTTCA | 105 |
| LlDEAD43-R | TTCCAGGGGTAGCCACAA | |
| LlDEAD65-F | CGGGCAGATTGGTGGATT | 165 |
| LlDEAD65-R | TGGTCTGTCTTTCACCTCGG |
Tab. 4
Physicochemical property and subcellular localization"
| 理化指标 | 蛋白质 | |||
|---|---|---|---|---|
| LlDEAD12 | LlDEAD32 | LlDEAD43 | LlDEAD65 | |
| 氨基酸的数量 | 1546 | 2619 | 2144 | 3302 |
| 分子量/Da | 129345.23 | 220714.18 | 176455.64 | 276389.07 |
| 等电点 | 4.94 | 4.82 | 4.94 | 4.82 |
| 稳定指数 | 56.86 | 51.35 | 38.34 | 47.32 |
| 脂肪指数 | 27.04 | 31.54 | 29.24 | 31.8 |
| 亲水性总平均值 | 0.915 | 0.882 | 0.711 | 0.864 |
| 原子总数 | 16283 | 28285 | 22738 | 35465 |
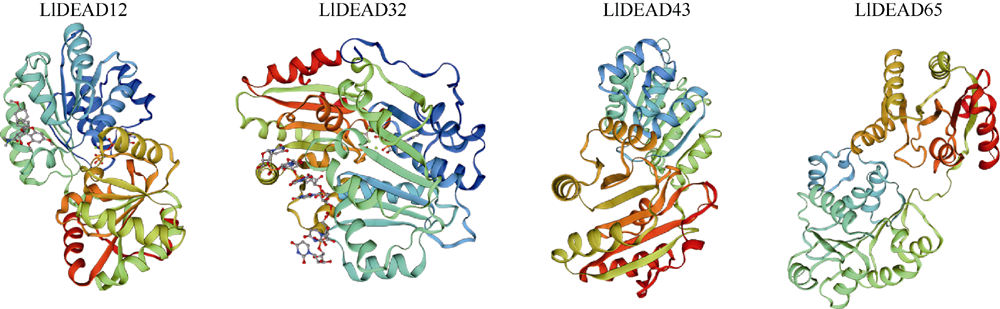
| 亚细胞定位 | 细胞质 | 细胞核 | 线粒体 | 线粒体 |

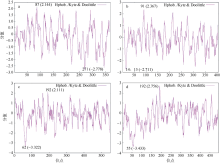
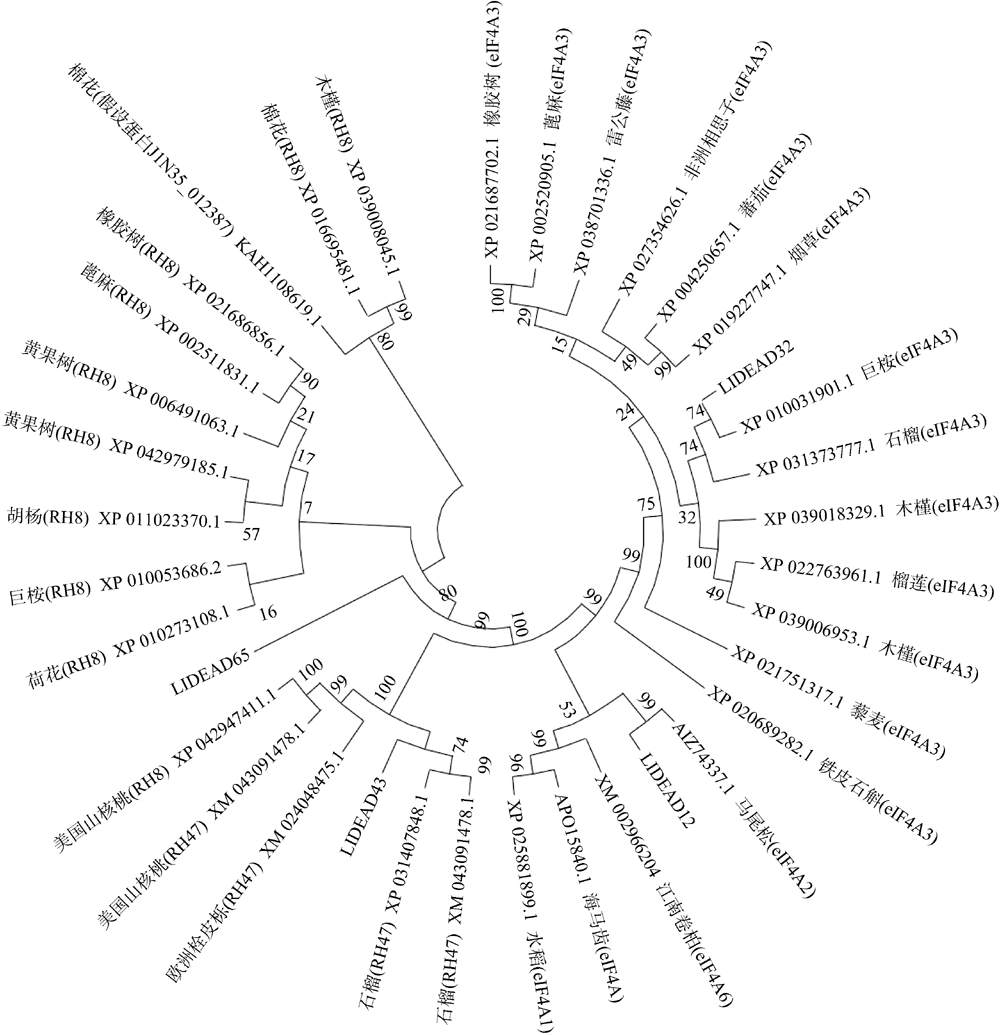
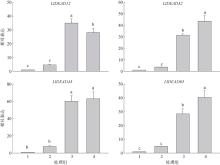
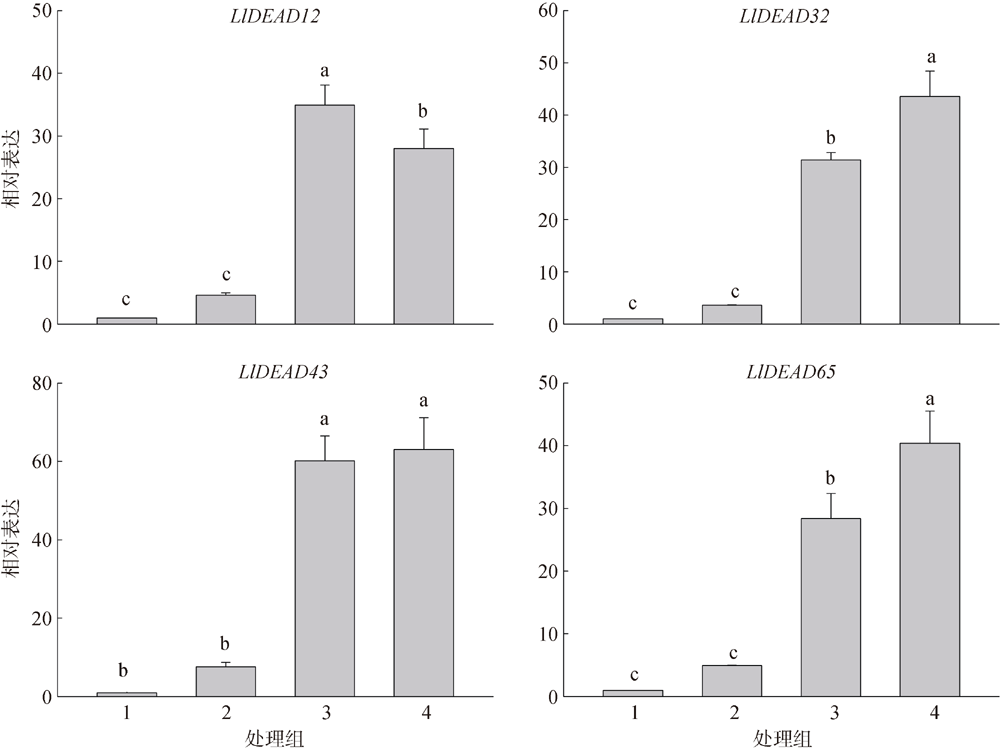
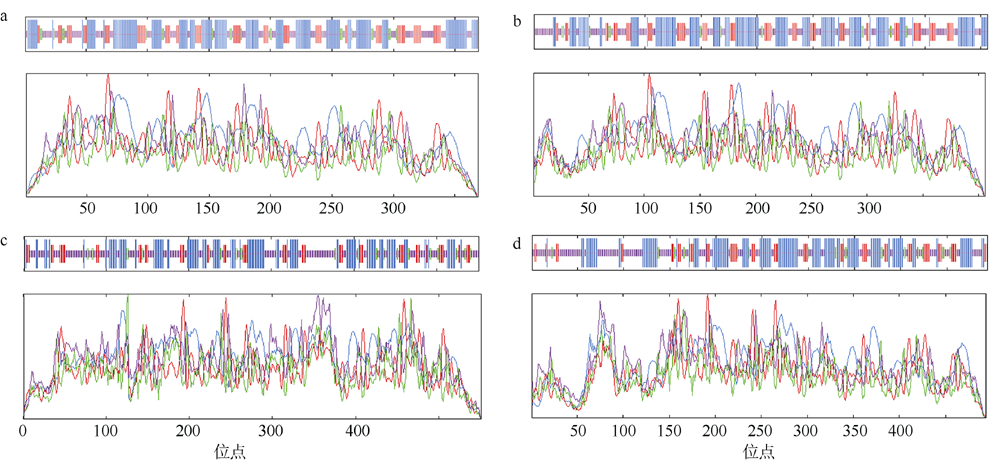
Fig. 2
Predicted secondary structure of DEAD-box protein. (a) LlDEAD12; (b) LlDEAD32; (c) LlDEA43; (d) LlDEAD65. Blue squares and lines in the figure represent α-helices; green squares and lines represent β-sheets; yellow squares and lines represent random coils; red squares and lines represent extended chains"
| [1] | 蔡敬, 孟小庆, 董婷婷, 等, 2017. DEAD-box解旋酶在植物非生物胁迫响应中的功能研究进展[J]. 生命科学, 29(5): 427-433. |
| CAI JING, MENG XIAOQING, DONG TINGTING, et al, 2017. Progress of plant DEAD-box helicase in response to abiotic stress[J]. Chinese Bulletin of Life Sciences, 29(5): 427-433. (in Chinese with English abstract) | |
| [2] | 陈丹, 彭雄波, 2016. DEAD-box基因家族在拟南芥生长发育中的作用[J]. 植物科学学报, 34(6): 941-948. |
| CHEN DAN, PENG XIONGBO, 2016. DEAD-box: function on growth and development in Arabidopsis thaliana[J]. Plant Science Journal, 34(6): 941-948. (in Chinese with English abstract) | |
| [3] | 孔荣荣, 2019. 水稻DEAD-box RNA解旋酶蛋白基因TCD33功能研究[D]. 上海: 上海师范大学. |
| KONG RONGRONG, 2019. Functional research of a DEAD-box RNA helicase gene TCD33 in rice (Oryza sativa L.)[D]. Shanghai: Shanghai Normal University. (in Chinese with English abstract) | |
| [4] | 铁原毓, 田洁, 2021. 大蒜蔗糖转化酶基因AsINV的克隆及其响应低温和干旱胁迫的表达分析[J]. 植物生理学报, 57(12): 2258-2270. |
| TIE YUANYU, TIAN JIE, 2021. Cloning of Allium sativum invertase gene AsINV and its expression analysis in response to low temperature and drought stress[J]. Plant Physiology Journal, 57(12): 2258-2270. (in Chinese with English abstract) | |
| [5] | 张颖, 陈光程, 钟才荣, 2021. 中国濒危红树植物研究与恢复现状[J]. 应用海洋学学报, 40(1): 142-153. |
| ZHANG YING, CHEN GUANGCHENG, ZHONG CAIRONG, 2021. Research on endangered mangrove species and recovery status in China[J]. Journal of Applied Oceanography, 40(1): 142-153.. (in Chinese with English abstract) | |
| [6] | 张颖, 钟才荣, 杨勇, 等, 2018. 濒危红树植物红榄李种质资源挽救[J]. 分子植物育种, 16(12): 4112-4118. |
| ZHANG YING, ZHONG CAIRONG, YANG YONG, et al, 2018. Rescue of germplasm resources of endangered mangrove plant Lumnitzera littorea[J]. Molecular Plant Breeding, 16(12): 4112-4118. (in Chinese with English abstract) | |
| [7] | 钟才荣, 李诗川, 管伟, 等, 2011. 中国3种濒危红树植物的分布现状[J]. 生态科学, 30(4): 431-435. |
| ZHONG CAIRONG, LI SHICHUAN, GUAN WEI, et al, 2011. Current distributions of three endangered mangrove species in China[J]. Ecological Science, 30(4): 431-435. (in Chinese with English abstract) | |
| [8] |
ANANTHARAMAN V, KOONIN E V, ARAVIND L, 2002. Comparative genomics and evolution of proteins involved in RNA metabolism[J]. Nucleic Acids Research, 30(7): 1427-1464.
pmid: 11917006 |
| [9] |
ANDERSON P, KEDERSHA N, 2006. RNA granules[J]. Journal of Cell Biology, 172(6): 803-808.
pmid: 16520386 |
| [10] |
ASAKURA Y, GALARNEAU E, WATKINS K P, et al, 2012. Chloroplast RH3 DEAD box RNA helicases in Maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis[J]. Plant Physiology, 159(3): 961-974.
doi: 10.1104/pp.112.197525 pmid: 22576849 |
| [11] |
BRAUD C, ZHENG WENGUANG, XIAO WENYAN, 2012. LONO1 encoding a nucleoporin is required for embryogenesis and seed viability in Arabidopsis[J]. Plant Physiology, 160(2): 823-836.
doi: 10.1104/pp.112.202192 |
| [12] |
CHEN PENGYUN, JIAN HONGLIANG, WEI FEI, et al, 2021. Phylogenetic analysis of the membrane attack complex/ perforin domain-containing proteins in Gossypium and the role of GhMACPF26 in cotton under cold stress[J]. Frontiers in Plant Science, 12: 684227.
doi: 10.3389/fpls.2021.684227 |
| [13] |
CHI WEI, HE BAOYE, MAO JUAN, et al, 2012. The function of RH22, a DEAD RNA helicase, in the biogenesis of the 50S ribosomal subunits of Arabidopsis chloroplasts[J]. Plant Physiology, 158(2): 693-707.
doi: 10.1104/pp.111.186775 pmid: 22170977 |
| [14] |
CORDIN O, BANROQUES J, TANNER N K, et al, 2006. The DEAD-box protein family of RNA helicases[J]. Gene, 367: 17-37.
pmid: 16337753 |
| [15] |
FAIRMAN-WILLIAMS M E, GUENTHER U P, JANKOWSKY E, 2010. SF1 and SF2 helicases: family matters[J]. Current Opinion in Structural Biology, 20(3): 313-324.
doi: 10.1016/j.sbi.2010.03.011 |
| [16] |
GE SHASHA, DUO LAN, WANG JUNQI, et al, 2021. A unique understanding of traditional medicine of pomegranate, Punica granatum L. and its current research status[J]. Journal of Ethnopharmacology, 271: 113877.
doi: 10.1016/j.jep.2021.113877 |
| [17] |
GOLLAN P J, TIKKANEN M, ARO E M, 2015. Photosynthetic light reactions: integral to chloroplast retrograde signalling[J]. Current Opinion in Plant Biology, 27: 180-191.
doi: 10.1016/j.pbi.2015.07.006 pmid: 26318477 |
| [18] |
GONG ZHIZHONG, DONG CHUNHAI, LEE H, et al, 2005. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis[J]. The Plant Cell, 17(1): 256-267.
doi: 10.1105/tpc.104.027557 |
| [19] |
JACOBSEN S E, RUNNING M P, MEYEROWITZ E M, 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems[J]. Development, 126(23): 5231-5243.
doi: 10.1242/dev.126.23.5231 pmid: 10556049 |
| [20] |
KIM J S, KIM K A, OH T R, et al, 2008. Functional characterization of DEAD-box RNA helicases in Arabidopsis thaliana under abiotic stress conditions[J]. Plant Cell Physiol, 49(10): 1563-71.
doi: 10.1093/pcp/pcn125 pmid: 18725370 |
| [21] |
KÖHLER D, SCHMIDT-GATTUNG S, BINDER, 2010. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana[J]. Plant Mol Biol, 72(4-5): 459-467.
doi: 10.1007/s11103-009-9584-9 |
| [22] |
KREPS JA, WU Y, CHANG HS, ZHU T, et al, 2002. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress[J]. Plant Physiol, 130(4): 2129-2141.
pmid: 12481097 |
| [23] |
KUMAR S, STECHER G, LI M, et al, 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol, 35(6): 1547-1549.
doi: 10.1093/molbev/msy096 pmid: 29722887 |
| [24] |
LI D, LIU H, ZHANG H, et al, 2008. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress[J]. J Exp Bot, 59(8): 2133-2146.
doi: 10.1093/jxb/ern072 pmid: 18441339 |
| [25] |
LI S C, CHUNG M C, CHEN C S, 2001. Cloning and characterization of a DEAD box RNA helicase from the viable seedlings of aged mung bean[J]. Plant Mol Biol, 47(6): 761-770.
doi: 10.1023/a:1013687412020 pmid: 11785937 |
| [26] | LINDER P, JANKOWSKY E, 2011. From unwinding to clamping the DEAD box RNA helicase family[J]. Nat Rev Mol Cell Biol, 12(8): 505-516. |
| [27] |
LINDER P, 2006. Dead-box proteins: a family affairactive and passive players in RNP-remodeling[J]. Nucleic Acids Res, 34(15): 4168-80.
doi: 10.1093/nar/gkl468 |
| [28] |
LIU Y, TABATA D, IMAI R, 2016. A cold-inducible DEAD-Box RNA Helicase from Arabidopsis thaliana regulates plant growth and development under low temperature[J]. PLoS One, 11(4): e0154040.
doi: 10.1371/journal.pone.0154040 |
| [29] |
LIVAK K J, SCHMITTGEN T D, 2001. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2-ΔΔCT method[J]. Methods, 25(4): 402-408.
doi: 10.1006/meth.2001.1262 |
| [30] |
LORKOVIC ZJ, HERMANN RG, OELMULLER R, 1997. PRH75, a new nucleus-localized member of the DEAD-box protein family from higher plants[J]. Mol Cell Biol, 17(4): 2257-2265.
doi: 10.1128/MCB.17.4.2257 pmid: 9121476 |
| [31] |
LUO Y, LIU YB, DONG YX, et al, 2009. Expression of a putative alfalfa helicase increases tolerance to abiotic stress in Arabidopsis by enhancing the capacities for ROS scavenging and osmotic adjustment[J]. J Plant Physiol, 166(4): 385-394.
doi: 10.1016/j.jplph.2008.06.018 |
| [32] |
MATTES A, SCHMIDT-GATTUND S, KOHLER D, et al, 2007. Two DEAD-box proteins may be part of RNA-dependent high-molecular-mass protein complexes in Arabidopsis mitochondria[J]. Plant Physiol, 145(4): 1637-1646.
doi: 10.1104/pp.107.108076 pmid: 17951454 |
| [33] |
MAZROUI R, SUKARIEH R, BORDELEAU ME, et al, 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation[J]. Mol Biol Cell, 17(10): 4212-4219.
pmid: 16870703 |
| [34] |
MATTHES A, SCHMIDT-GATTUNG S, KOHLER D, et al, 2007. Two DEAD-box proteins may be part of RNA-dependent high-molecularmass protein complexes in Arabidopsis mitochondria[J]. Plant Physiol, 145(4): 1637-1646.
doi: 10.1104/pp.107.108076 |
| [35] |
NAWAZ G, LEE K, PARK S J, et al, 2018a. A chloroplast-targeted cabbage DEAD-box RNA helicase BrRH22 confers abiotic stress tolerance to transgenic Arabidopsis plants by affecting translation of chloroplast transcripts[J]. Plant Physiology and Biochemistry, 127: 336-342.
doi: 10.1016/j.plaphy.2018.04.007 |
| [36] |
NAWAZ G, SAI T Z T, LEE K, et al, 2018b. Rice DEAD-box RNA helicase OsRH53 has negative impact on Arabidopsis response to abiotic stresses[J]. Plant Growth Regulation, 85(1): 153-163.
doi: 10.1007/s10725-018-0381-9 |
| [37] |
NAWAZ G, KANG H, 2019. Rice OsRH58, a chloroplast DEAD-box RNA helicase, improves salt or drought stress tolerance in Arabidopsis by affecting chloroplast translation[J]. BMC Plant Biol, 19(1): 17.
doi: 10.1186/s12870-018-1623-8 |
| [38] |
NIDUMUKKALA S, TAYI L, CHITTELA R K, et al, 2019. DEAD box helicases as promising molecular tools for engineering abiotic stress tolerance in plants[J]. Critical Reviews in Biotechnology, 39(3): 395-407.
doi: 10.1080/07388551.2019.1566204 pmid: 30714414 |
| [39] |
NISHIMURA K, ASHIDA H, OGAWA T, et al, 2010. A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA[J]. The Plant Journal, 63(5): 766-777.
doi: 10.1111/j.1365-313X.2010.04276.x pmid: 20561259 |
| [40] |
OBERSCHELP G P J, GUARNASCHELLI A B, TESON N, et al, 2020. Cold acclimation and freezing tolerance in three Eucalyptus species: a metabolomic and proteomic approach[J]. Plant Physiology and Biochemistry, 154: 316-327.
doi: 10.1016/j.plaphy.2020.05.026 |
| [41] |
OP DEN CAMP R, KUHLEMEIER C, 1998. Phosphorylation of tobacco eukaryotic translation initiation factor 4A upon pollen tube germination[J]. Nucleic Acids Research, 26(9): 2058-2062.
pmid: 9547259 |
| [42] |
REN QIFEI, ZHOU YUNCHAO, ZHOU XINWEI, 2020. Combined transcriptome and proteome analysis of Masson Pine (Pinus massoniana Lamb.) seedling root in response to nitrate and ammonium supplementations[J]. International Journal of Molecular Sciences, 21(2): 7548.
doi: 10.3390/ijms21207548 |
| [43] |
SEKI M, NARUSAKA M, ABE H, et al, 2001. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray[J]. The Plant Cell, 13(1): 61-72.
doi: 10.1105/tpc.13.1.61 |
| [44] |
STONEBLOOM S, BURCH-SMITH T, KIM I, et al, 2009. Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata[J]. Proceedings of the National Academy of Sciences of the United States of America, 106(40): 17229-17234.
doi: 10.1073/pnas.0909229106 pmid: 19805190 |
| [45] |
TANNER N K, LINDER P, 2001. DExD/H box RNA helicases[J]. Molecular Cell, 8(2): 251-262.
doi: 10.1016/S1097-2765(01)00329-X |
| [46] | TUTEJA N, SAHOO R K, GARG B, et al, 2013. OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64)[J]. The Plant Journal, 76(1): 115-127. |
| [47] |
VASHISHT A A, PRADHAN A, TUTEJA R, et al, 2005. Cold- and salinity stress-induced bipolar pea DNA helicase 47 is involved in protein synthesis and stimulated by phosphorylation with protein kinase C[J]. The Plant Journal, 44(1): 76-87.
doi: 10.1111/j.1365-313X.2005.02511.x |
| [48] |
VASHISHT A A, TUTEJA N, 2006. Stress responsive DEAD-box helicases: a new pathway to engineer plant stress tolerance[J]. Journal of Photochemistry and Photobiology B: Biology, 84(2): 150-160.
doi: 10.1016/j.jphotobiol.2006.02.010 |
| [49] |
WEBSTER C, GAUT R L, BROWNING K S, et al, 1991. Hypoxia enhances phosphorylation of eukaryotic initiation factor 4A in maize root tips[J]. Journal of Biological Chemistry, 266(34): 23341-23346.
pmid: 1744128 |
| [50] |
WESTERN T L, CHENG YULAN, LIU JUN, et al, 2002. HUA ENHANCER2, a putative DExH-box RNA helicase, maintains homeotic B and C gene expression in Arabidopsis[J]. Development, 129(7): 1569-1581.
doi: 10.1242/dev.129.7.1569 |
| [51] |
ZHANG BIN, JIA DONG, GAO ZHIQIANG, et al, 2016. Physiological responses to low temperature in spring and winter wheat varieties[J]. Journal of the Science of Food and Agriculture, 96(6): 1967-1973.
doi: 10.1002/jsfa.7306 pmid: 26095741 |
| [52] |
ZHU MINGKU, CHEN GUOPING, DONG TINGTING, et al, 2015. SlDEAD31, a putative DEAD-box RNA helicase gene, regulates salt and drought tolerance and stress-related genes in tomato[J]. PLoS One, 10(8): e0133849.
doi: 10.1371/journal.pone.0133849 |
| [1] | AN Fan, JIANG Yue, WANG Yu, CAO Guangping, GAO Chenghai, LIU Yonghong, YI Xiangxi, BAI Meng. Studies on secondary metabolites of endophytic fungus Aspergillus terreus GXIMD 03158 isolated from mangroves Acanthus ilicifolius L. [J]. Journal of Tropical Oceanography, 2024, 43(5): 41-48. |
| [2] | WU Hongbo, LUO Feng, CHEN Zhipeng, ZHU Fei, ZENG Jingwei, ZHANG Chi, LI Ruijie. A novel pattern for predicting the effects of mangrove ecological reconstruction [J]. Journal of Tropical Oceanography, 2024, 43(4): 86-97. |
| [3] | ZHENG Fa, HUANG Fulin, CHEN Zeheng, DING Weipin. Mangrove wetland dynamics in Shankou, Guangxi based on LUCC and landscape pattern change [J]. Journal of Tropical Oceanography, 2024, 43(4): 165-173. |
| [4] | ZHOU Zhigang, YUE Wen, LI Huiquan, LIN Yangyang. Influence of tree species and intertidal elevations on the carbon storage of the Gaoqiao mangrove area in Zhanjiang, Guangdong Province [J]. Journal of Tropical Oceanography, 2024, 43(2): 108-120. |
| [5] | SHEN Jian, JIAN Zhuokai, OUYANG Xuemin, AI Bin. Remote sensing monitoring of mangrove wetland changes combined with tidal level correction in the Leizhou Peninsula [J]. Journal of Tropical Oceanography, 2024, 43(1): 137-153. |
| [6] | GENG Wanlu, XING Yongze, ZHANG Qiufeng, GUAN Weibing. Structural characteristics of macrobenthic communities at intertidal zone for mangrove in Beihai, Guangxi [J]. Journal of Tropical Oceanography, 2024, 43(1): 107-115. |
| [7] | DONG Junde, HUANG Xiaofang, LONG Aimin, WANG Youshao, LING Juan, YANG Qingsong. Progress on the nitrogen-fixing microorganisms and their ecological functions in mangroves [J]. Journal of Tropical Oceanography, 2023, 42(4): 1-11. |
| [8] | LIANG Hanqiao, CHEN Wenfeng, FAN Yikai, ZHU Zidong, MA Guoxu, CHEN Deli, TIAN Jing. Research progress on the secondary metabolites and activities of endophytic fungi of genus Aspergillus and Trichoderma from mangroves [J]. Journal of Tropical Oceanography, 2023, 42(4): 12-24. |
| [9] | QIU Jin, DAI Hongtao, XING Yongze, HUANG Daji, Yin Qunjian, CHENG Dewei. Leaves, stems and roots stoichiometry characteristics of mangrove plants at different succession stages in the Shankou National Mangrove Nature Reserve, China* [J]. Journal of Tropical Oceanography, 2023, 42(3): 149-157. |
| [10] | ZHANG Chengfei, REN Guangbo, WU Peiqiang, HU Yabin, MA Yi, YAN Yu, ZHANG Jingrui. Mangrove species classification in the Hainan Bamen Bay based on GF optics and fully polarimetric SAR [J]. Journal of Tropical Oceanography, 2023, 42(2): 153-168. |
| [11] | YE Jincheng, CHEN Yiqing, GAO Lin, ZHOU Xianjiao, ZHONG Cairong, ZHANG Ying, WANG Yun. Analysis of rhizosphere bacterial community characteristics of mangrove plant Sonneratia × gulngai and its parents [J]. Journal of Tropical Oceanography, 2022, 41(6): 75-89. |
| [12] | SU Boyu, ZHANG Weishi, WANG Youshao. Response of antioxidant enzyme systems in root tissues of three mangrove species to waterlogging stress [J]. Journal of Tropical Oceanography, 2022, 41(6): 35-43. |
| [13] | INYANG Aniefiok Ini, ZHOU Yueyue, WANG Youshao. Characteristics of water quality and their eutrophication assessment on the mangrove ecosystems along the Guangdong coast [J]. Journal of Tropical Oceanography, 2022, 41(6): 1-11. |
| [14] | WU Weizhi, ZHAO Zhixia, YANG Sheng, LIANG Licheng, Chen Qiuxia, LU Xiang, LIU Xing, ZHANG Xiaowei. The mangrove forest distribution and analysis of afforestation effect in Zhejiang Province [J]. Journal of Tropical Oceanography, 2022, 41(6): 67-74. |
| [15] | XIE Yong, WANG Youshao, ZHANG Weishi. Cloning and expression analysis of Cinnamate-4-hydroxylase gene from Rhizophora stylosa [J]. Journal of Tropical Oceanography, 2022, 41(6): 20-27. |
|
||