Journal of Tropical Oceanography ›› 2019, Vol. 38 ›› Issue (6): 80-89.doi: 10.11978/2019012CSTR: 32234.14.2019012
• Marine Biology • Previous Articles Next Articles
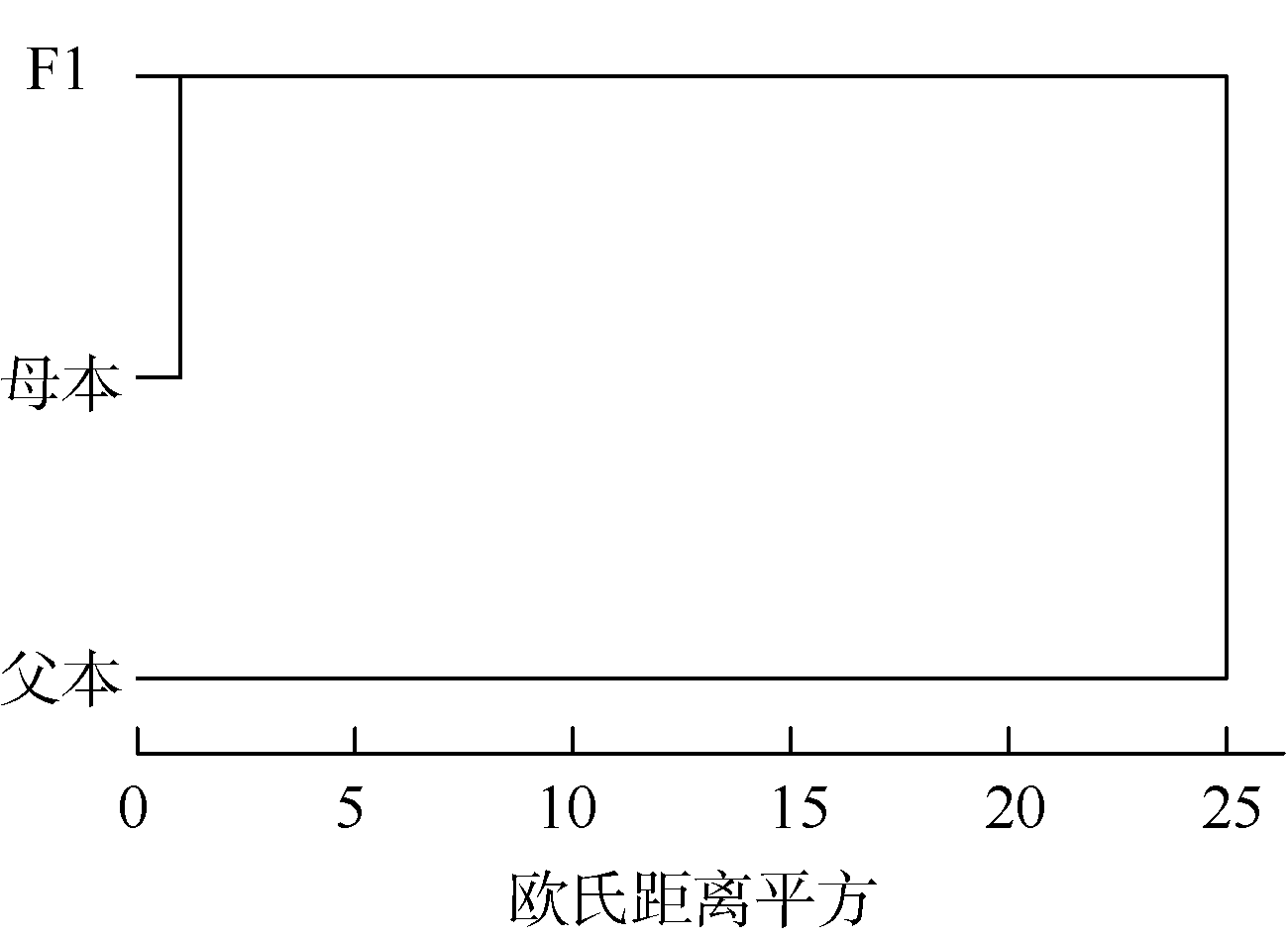
Morphological and SNP markers for analysis of genetic structure of hybrid progeny and their parental populations of Pinctada fucata martensii
HUANG Jing1,2,3, PAN Xiaolan1,2,3, XU Meng1,2,3, LIU Wenguang1,3, ZHANG Hua1,3, HE Maoxian1,3( )
)
- 1. Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
2. University of Chinese Academy of Sciences, Beijing 100049, China
3. Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
-
Received:2019-01-21Revised:2019-02-27Online:2019-11-20Published:2019-11-26 -
Contact:Maoxian HE E-mail:hmx2@scsio.ac.cn -
Supported by:Modern Agro-industry Technology Research System(CARS-49)
Cite this article
HUANG Jing, PAN Xiaolan, XU Meng, LIU Wenguang, ZHANG Hua, HE Maoxian. Morphological and SNP markers for analysis of genetic structure of hybrid progeny and their parental populations of Pinctada fucata martensii[J].Journal of Tropical Oceanography, 2019, 38(6): 80-89.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
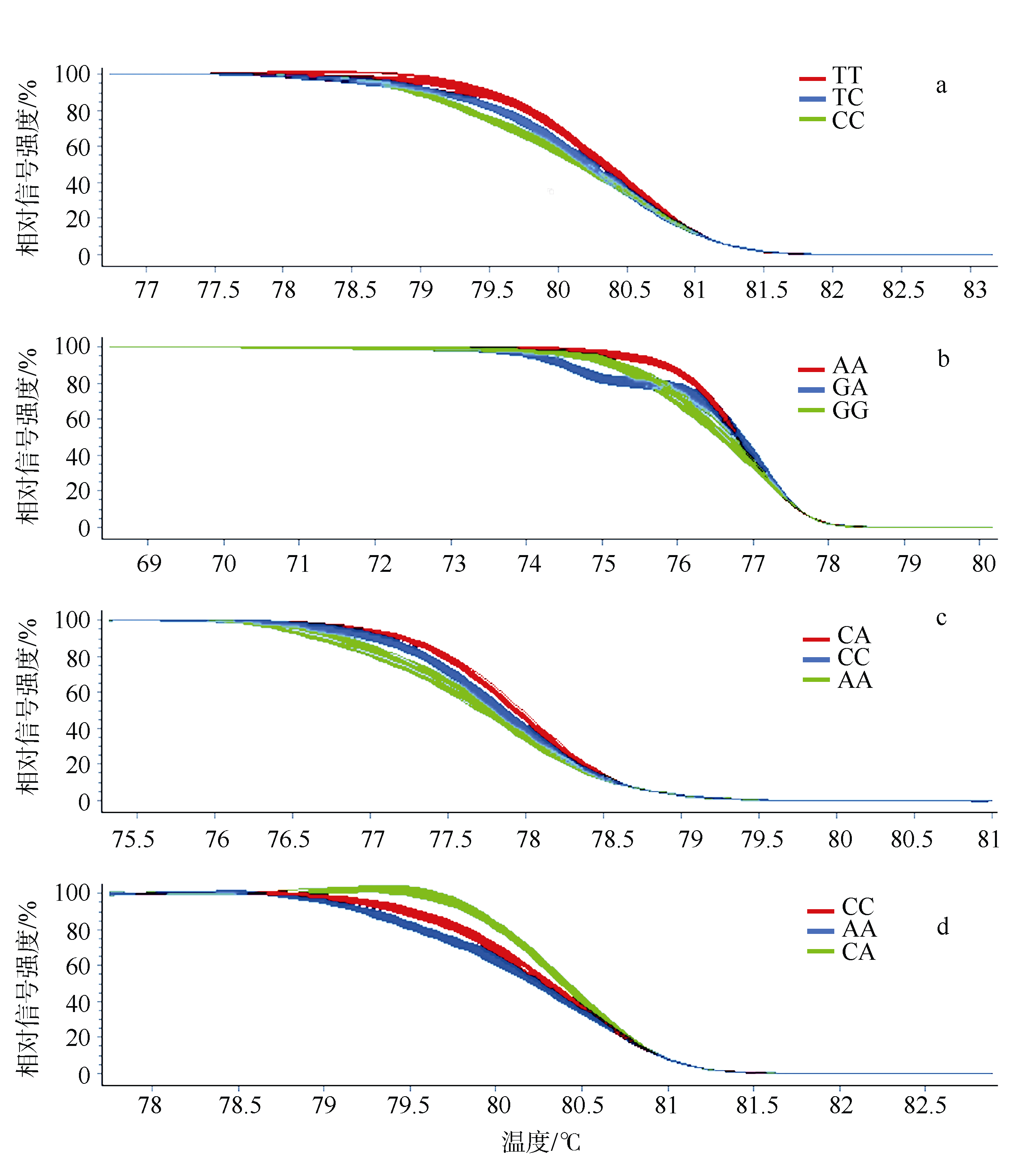
Primers used for gene genotyping by HRM"
| 位点名称 | 序列(5'-3') | SNP 类型 | 退火温度/℃ | 对应产物长度/bp |
|---|---|---|---|---|
| rs8 | F: CCCAGCATACTTTATGGCAGAAT R: CCTAGCGTGGAGAAGAGATGTA | T/C | 55 | 122 |
| rs33 | F: CAATACTCTCAGGGACATCTT R: CTCAACAGTGTCGCTTATATG | A/G | 50 | 107 |
| rs35 | F: TTCCGCAGGATGAATTTAGGT R: TCTGAAGTCTGTTTGTCCAATG | C/A | 53 | 103 |
| rs44 | F: CCAAAGCCAACTACACCCAAA R: GTCAAACGCCCACGATACAC | C/A | 55 | 144 |
Tab. 2
Average values of main morphological parameters in three populations of P. fucata martensii"
| 性状 | 父本 | F1 | 母本 |
|---|---|---|---|
| 壳长 | 58.25±5.58a | 56.40±5.20a | 56.52±4.29a |
| 壳宽 | 23.44±2.23a | 20.23±2.84b | 21.72±2.43ab |
| 壳高 | 59.24±5.10a | 55.60±5.47b | 56.25±4.45b |
| 铰合线 | 51.16±3.79a | 50.17±4.97a | 50.84±4.28a |
| 总重 | 28.16±6.07a | 24.60±7.49b | 24.12±5.33b |
| 壳重 | 14.23±2.78a | 13.11±3.78ab | 11.95±2.62b |
| 软体部重 | 10.77±3.25a | 8.81±3.38b | 8.49±2.64b |
| 壳宽指数 | 0.17±0.01a | 0.15±0.01b | 0.16±0.01ab |
| 壳重指数 | 18.12±2.75b | 20.42±2.06a | 17.43±2.60b |
| 肥满度指数 | 13.36±2.27a | 13.28±2.53a | 13.02±2.25a |
| 壳高/壳长 | 1.02±0.05a | 0.99±0.05b | 1.00±0.06ab |
| 壳宽/壳长 | 0.41±0.03a | 0.36±0.03b | 0.38±0.03ab |
| 铰合线/壳长 | 0.88±0.05b | 0.89±0.05ab | 0.90±0.05a |
Tab. 3
Principal component analysis of three populations of P. fucata martensii by morphological parameters"
| 性状 | 负荷值 | ||
|---|---|---|---|
| 主成分1 | 主成分2 | 主成分3 | |
| 总重 | 0.969* | 0.019 | -0.040 |
| 软体部重 | 0.954* | 0.021 | 0.107 |
| 壳高 | 0.937* | -0.052 | -0.030 |
| 壳重 | 0.932* | -0.027 | -0.103 |
| 铰合线长 | 0.901* | 0.064 | 0.069 |
| 壳长 | 0.884* | 0.067 | -0.433 |
| 壳宽指数 | 0.113 | 0.952* | 0.173 |
| 壳宽 | 0.301 | 0.938* | 0.061 |
| 壳宽/壳长 | 0.102 | 0.936* | 0.250 |
| 壳重指数 | 0.184 | -0.758 | 0.088 |
| 肥满度 | 0.454 | -0.531 | 0.393 |
| 铰合线/壳长 | 0.089 | 0.003 | 0.846* |
| 壳高壳/长 | 0.250 | -0.238 | 0.688* |
| 贡献率/% | 43.2 | 27.6 | 12.8 |
Tab. 5
Genetic diversity parameters of three populations of P. fucata martensii"
| 群体名称 | 位点 | I | Ho | He | PIC | HWE P值 |
|---|---|---|---|---|---|---|
| 父本 | rs8 | 0.3768 | 0.2500 | 0.2222 | 0.1948 | 0.4527 |
| rs33 | 0.6809 | 0.3438 | 0.4955 | 0.3685 | 0.0780 | |
| rs35 | 0.3111 | 0.0625 | 0.1726 | 0.1555 | 0.0044* | |
| rs44 | 0.6211 | 0.1875 | 0.4365 | 0.3384 | 0.0010* | |
| 平均值 | 0.4975 | 0.2110 | 0.3317 | 0.2643 | ||
| F1 | rs8 | 0.6002 | 0.1515 | 0.4163 | 0.3260 | 0.0003* |
| rs33 | 0.6816 | 0.3030 | 0.4960 | 0.3692 | 0.0230 | |
| rs35 | 0.5860 | 0.4242 | 0.4028 | 0.3182 | 0.7500 | |
| rs44 | 0.6635 | 0.2727 | 0.4779 | 0.3598 | 0.0120 | |
| 平均值 | 0.6328 | 0.2879 | 0.4483 | 0.3433 | ||
| 母本 | rs8 | 0.6881 | 0.4333 | 0.5034 | 0.3724 | 0.4380 |
| rs33 | 0.5799 | 0.3333 | 0.3977 | 0.3137 | 0.3622 | |
| rs35 | 0.6657 | 0.0333 | 0.4808 | 0.3614 | 0.0000* | |
| rs44 | 0.6926 | 0.2333 | 0.4921 | 0.3747 | 0.0025* | |
| 平均值 | 0.6566 | 0.2583 | 0.4685 | 0.3556 |
Tab. 6
Genotypes and frequencies of SNP in three populations of P. fucata martensii"
| 群体名称 | 基因型(频率) | |||
|---|---|---|---|---|
| rs8 | rs33 | rs35 | rs44 | |
| 父本 | TT(0.75) | GG(0.41) | CC(0.88) | CC(0.59) |
| TC(0.25) | GA(0.34) | CA(0.06) | CA(0.19) | |
| CC(0) | AA(0.25) | AA(0.06) | AA(0.22) | |
| F1 | TT(0.64) | GG(0.27) | CC(0.52) | CC(0.24) |
| TC(0.15) | GA(0.30) | CA(0.42) | CA(0.27) | |
| CC(0.21) | AA(0.42) | AA(0.06) | AA(0.49) | |
| 母本 | TT(0.23) | GG(0.10) | CC(0.60) | CC(0.37) |
| TC(0.43) | GA(0.33) | CA(0.03) | CA(0.23) | |
| CC(0.33) | AA(0.57) | AA(0.37) | AA(0.40) | |
| 1 | 白牡丹, 王彩虹, 殷豪 , 等, 2012. 苹果不同HRM反应体系分析效果评价[J]. 分子植物育种, 10(1):115-120. |
| BAI MUDAN, WANG CAIHONG, YIN HAO , et al, 2012. Evaluation of different reaction systems for HRM analysis in apple[J]. Molecular Plant Breeding, 10(1):115-120 (in Chinese with English abstract). | |
| 2 | 陈亮, 赵柏淞, 李玲 , 等, 2014. 人工诱导栉孔扇贝雌核发育胚胎的SNP标记分析[J]. 中国海洋大学学报, 44(2):48-52. |
| CHEN LIANG, ZHAO BOSONG, LI LING , et al, 2014. Genotyping artificially induced gynogenetic embryos of Zhikong scallop (Chlamys farreri) with SNP markers[J]. Periodical of Ocean University of China, 44(2):48-52 (in Chinese with English abstract). | |
| 3 | 陈蓉, 刘建勇, 唐连俊 , 等, 2009. 中国5个海区不同群体毛蚶形态差异分析[J]. 海洋科学, 33(7):64-69. |
| CHEN RONG, LIU JIANYONG, TANG LIANJUN , et al, 2009. Morphological variations analysis of five different populations of Scapharca subcrenata in China[J]. Marine Sciences, 33(7):64-69 (in Chinese with English abstract). | |
| 4 | 董志国, 李家乐, 郑汉丰 , 2008. 三角帆蚌3个地理种群自交与杂交F1代的形态差异分析[J]. 大连水产学院学报, 23(2):92-97. |
| DONG ZHIGUO, LI JIALE, ZHENG HANFENG , 2008. Morphological variability in original parents and their reciprocal hybrids F1 of fresh water mussel Hyriopsis cumingii from three geographical populations in China[J]. Journal of Dalian Fisheries University, 23(2):92-97 (in Chinese with English abstract). | |
| 5 | 杜晓东, 李广丽, 刘志刚 , 等, 2002. 合浦珠母贝2个野生种群的遗传多样性[J]. 中国水产科学, 9(2):100-105. |
| DU XIAODONG, LI GUANGLI, LIU ZHIGANG , et al, 2002. Genetic diversity of two wild populations in Pinctada martensii[J]. Journal of Fishery Sciences of China, 9(2):100-105 (in Chinese with English abstract). | |
| 6 | 谷龙春, 李金碧, 喻达辉 , 等, 2010. 合浦珠母贝双列杂交家系的建立与遗传分析[J]. 水产学报, 34(1):26-31. |
| GU LONGCHUN, LI JINBI, YU DAHUI , et al, 2010. Establishment and genetic analysis of complete diallel cross families of pearl oyster (Pinctada fucata)[J]. Journal of Fisheries of China, 34(1):26-31 (in Chinese with English abstract). | |
| 7 | 顾志峰, 王嫣, 石耀华 , 等, 2009. 马氏珠母贝两个不同地理种群的形态性状和贝壳珍珠质颜色比较分析[J]. 渔业科学进展, 30(1):79-86. |
| GU ZHIFENG, WANG YAN, SHI YAOHUA , et al, 2009. Comparison of morphometrics and shell nacre colour between two geographical populations of pearl oyster Pinctada martensii (Dunker)[J]. Marine Fisheries Research, 30(1):79-86 (in Chinese with English abstract). | |
| 8 |
侯战辉, 王嫣, 石耀华 , 等, 2008. 马氏珠母贝(Pinctada martensii)2个不同地理种群遗传变异的EST-SSR分析[J]. 海洋与湖沼, 39(2):178-183.
doi: 10.11693/hyhz20080229029 |
|
HOU ZHANHUI, WANG YAN, SHI YAOHUA , et al, 2008. EST-SSR analysis of genetic variation between two geographical populations of pearl oyster, Pinctada martensii (Dunker)[J]. Oceanologia et Limnologia Sinica, 39(2):178-183 (in Chinese with English abstract).
doi: 10.11693/hyhz20080229029 |
|
| 9 | 胡笑蓉, 毛雄英, 王卫华 , 等, 2012. 一种基于HRM技术的快速DNA甲基化检测方法[J]. 中国卫生检验杂志, 22(6):1336-1338. |
| HU XIAORONG, MAO XIONGYING, WANG WEIHUA , et al, 2012. A novel and reliable method of DNA methylation analysis based on HRM technology[J]. Chinese Journal of Health Laboratory Technology, 22(6):1336-1338 (in Chinese with English abstract). | |
| 10 | 姜因萍, 何毛贤, 林岳光 , 2007. 马氏珠母贝群体内遗传多样性的ISSR分析[J]. 海洋通报, 26(5):62-66. |
| JIANG YINPING, HE MAOXIAN, LIN YUEGUANG , 2007. Genetic diversity of a cultured population on Pinctada Martensii Dunker by ISSR marker[J]. Marine Science Bulletin, 26(5):62-66 (in Chinese with English abstract). | |
| 11 | 金玉琳 , 2014. 长牡蛎SNP标记开发及其在家系分析和物种鉴定中的应用[D]. 青岛: 中国海洋大学: 32-38. |
| JIN YULIN , 2014. Development and characterization of SNP markers and their application in family analysis and species identification for Pancific oyster Crassostrea gigas[D]. Qingdao: Ocean University of China: 32-38 (in Chinese with English abstract). | |
| 12 | 李纪勤 , 2012. 栉孔扇贝(Chlamys farreri)EST-SNP的开发及其应用[D]. 青岛: 中国海洋大学: 53-54. |
| LI JIQIN , 2012. Development, characterization and application of EST-SNP markers in Zhikong scallop (Chlamys farreri)[D]. Qingdao: Ocean University of China: 53-54 (in Chinese with English abstract). | |
| 13 | 李梅, 吕伟朋, 韦鸿 , 等, 2012. HRM方法检测非小细胞肺癌患者胸水标本癌细胞基因突变的临床意义[J]. 大连医科大学学报, 34(4):321-323, 347. |
| LI MEI, LÜ WEIPENG, WEI HONG , et al, 2012. Feasibility of HRM to detect gene mutations in hydrothorax of patients with non -small cell lung cancer[J]. Journal of Dalian Medical University, 34(4):321-323, 347 (in Chinese with English abstract). | |
| 14 | 李太武, 孙修勤, 刘艳 , 等, 2002. 中日栉孔扇贝杂交子一代群体的遗传变异[J]. 高技术通讯, 12(6):101-105, 100. |
| LI TAIWU, SUN XIUQIN, LIU YAN , et al, 2002. Allozyme variation of two subspecies of Chlamys farreri and their reciprocal hybrids[J]. High Technology Letters, 12(6):101-105, 100 (in Chinese with English abstract). | |
| 15 | 李耀国, 刘文广, 林坚士 , 等, 2016. 马氏珠母贝SNP标记开发及家系遗传多态性分析[J]. 海洋通报, 35(1):96-102. |
| LI YAOGUO, LIU WENGUANG, LIN JIANSHI , et al, 2016. Development of SNP markers in Pinctada fucata and its application for family genetic analysis[J]. Marine Science Bulletin, 35(1):96-102 (in Chinese with English abstract). | |
| 16 | 刘泽浩, 李捷, 杨学玲 , 等, 2010 中国北方沿海潮间带常见镜蛤的种类[J]. 海洋科学, 34(12):30-35. |
| LIU ZEHAO, LI JIE, YANG XUELING , et al, 2010. Species of common Dosinia from intertidal zones of northern China[J]. Marine Sciences, 34(12):30-35 (in Chinese with English abstract). | |
| 17 | 罗会, 刘宝锁, 黎火金 , 等, 2013. 合浦珠母贝不同地理种群的形态差异和判别分析[J]. 广东农业科学, 40(12):171-174. |
| LUO HUI, LIU BAOSUO, LI HUOJIN , et al, 2013. Morphological variations and discriminant analysis on four populations of Pinctada fucata[J]. Guangdong Agricultural Sciences, 40(12):171-174 (in Chinese with English abstract). | |
| 18 | 吕林兰, 杜晓东, 王嫣 , 等, 2008. 马氏珠母贝3个野生种群及种群间杂交后代遗传多样性的ISSR分析[J]. 水生生物学报, 32(1):26-32. |
| LÜ LINLAN, DU XIAODONG, WANG YAN , et al, 2008. Genetic diversity of three popultions and the first generations of hybridization between different populations of pearl oyster, Pinctada martensii (Dunker)[J]. Acta Hydrobiologica Sinica, 32(1):26-32 (in Chinese with English abstract). | |
| 19 | 曲艳波, 叶翚, 石耀华 , 等, 2006. 马氏珠母贝杂交选育群体遗传变异的RAPD分析[C]// 海南生物技术研究与发展研讨会论文集. 三亚: 中国生物工程学会: 27-34. |
| QU YANBO, YE HUI, SHI YAOHUA , et al, 2006. RAPD analysis of genetic variation among cross breeding system of pearl oyster[C]// Proceedings of the Workshop on Research & Development of Biotechnology in Hainan. Sanya: Chinese Society of Biotechnology: 27-34 (in Chinese with English abstract). | |
| 20 | 沈恩健 , 2010. 利用AFLP和SSR对南海区域马氏珠母贝群体的遗传结构分析[D]. 湛江: 广东海洋大学. |
| SHEN ENJIAN , 2010. Genetic diversrty analyses of Pinctada fucata (dunker) in south China sea using AFLP and SSR[D]. Zhanjiang: Guangdong Ocean University (in Chinese with English abstract). | |
| 21 | 苏天凤, 蔡云川, 张殿昌 , 等, 2002. 合浦珠母贝3个养殖群体的RAPD分析[J]. 中国水产科学, 9(2):106-109. |
| SU TIANFENG, CAI YUNCHUAN, ZHANG DIANCHANG , et al, 2002. RAPD analysis of three cultured populations of Pinctada martensii[J]. Journal of Fishery Sciences of China, 9(2):106-109 (in Chinese with English abstract). | |
| 22 | 汤健, 管云雁, 刘文广 , 等, 2013. 马氏珠母贝家系遗传结构的微卫星分析[J]. 海洋科学, 37(8):35-41. |
| TANG JIAN, GUAN YUNYAN, LIU WENGUANG , et al, 2013. Microsatellite DNA analysis of genetic structures about 9 families of Pinctada fucata[J]. Marine Sciences, 37(8):35-41 (in Chinese with English abstract). | |
| 23 | 童晓飞, 郭莹, 丛晓霏 , 2012. 高分辨溶解曲线及其在检测海洋经济贝类SNPs的应用前景[J]. 北京农业, ( 15):139. |
| TONG XIAOFEI, GUO YING, CONG XIAOFEI , 2012. The high-resolution melting and the application foreground used in detecting SNPs of the biomonitor[J]. Beijing Agriculture, ( 15):139 (in Chinese with English abstract). | |
| 24 | 王爱民, 石耀华 , 2007. 马氏珠母贝的遗传改良技术[M] //王清印. 海水养殖生物的细胞工程育种. 北京: 海洋出版社: 133-164. |
| 25 |
王爱民, 王嫣, 顾志峰 , 等, 2010. 马氏珠母贝(Pinctada martensii)2个地理群体杂交子代的杂种优势和遗传变异[J]. 海洋与湖沼, 41(1):140-147.
doi: 10.11693/hyhz201001020020 |
|
WANG AIMIN, WANG YAN, GU ZHIFENG , et al, 2010. Heterosis and genetic variation of hybrids from two geographical populations of pearl oyster, Pinctada martensii[J]. Oceanologia et Limnologia Sinica, 41(1):140-147 (in Chinese with English abstract).
doi: 10.11693/hyhz201001020020 |
|
| 26 | 王家丰 , 2013. 长牡蛎基因区SNP标记规模开发及其在遗传育种研究中的应用[D]. 青岛: 中国科学院研究生院(海洋研究所). |
| WANG JIAFENG , 2013. Large-scale development of SNP marker and its potential application in genetic breeding of Crassostrea gigas[D]. Qingdao: Institute of Oceanology, Chinese Academy of Sciences (in Chinese with English abstract). | |
| 27 |
王艳, 赵雪, 姜振峰 , 等, 2015. 大豆小片段法HRM基因分型体系优化[J]. 中国油料作物学报, 37(4):453-461.
doi: 10.7505/j.issn.1007-9084.2015.04.004 |
|
WANG YAN, ZHAO XUE, JIANG ZHENFENG , et al, 2015. Optimization of small amplicon of HRM genotyping system in soybean[J]. Chinese Journal of Oil Crop Sciences, 37(4):453-461 (in Chinese with English abstract).
doi: 10.7505/j.issn.1007-9084.2015.04.004 |
|
| 28 | 杨润婷, 吴波, 李翀 , 等, 2013. 两种SNP分型方法的比较及其在柚品种鉴定中的应用[J]. 园艺学报, 40(6):1061-1070. |
| ZHAO RUNTING, WU BO, LI CHONG , et al, 2013. Comparison of allele-specific PCR and high resolution melting analysis in SNP genotyping and their application in Pummelo cultivar identification[J]. Acta Horticulturae Sinica, 40(6):1061-1070 (in Chinese with English abstract). | |
| 29 | 喻达辉, 李有宁, 吴开畅 . 2005. 中国、日本和澳大利亚珍珠贝的ITS2序列特征分析[J]. 南方水产, ( 02):4-9. |
| YU DAHUI, LI YOUNING, WU KAICHANG , 2005. Analysis on sequence variation of ITS 2 rDNA in Pinctada fucata from China, Japan and Australia[J]. South China Fisheries Science, 1(2):1-6 (in Chinese with English abstract). | |
| 30 | 张莉 , 2007. 中国珍珠产业的问题、困境与出路[J]. 农业现代化研究, 28(4):443-445. |
| ZHANG LI , 2007. Economical analysis of dilemma of Chinese pearl industry and outlet[J]. Research of Agricultural Modernization, 28(4):443-445 (in Chinese with English abstract). | |
| 31 | 赵杰, 游新勇, 徐贞贞 , 等, 2018. SNP检测方法在动物研究中的应用[J]. 农业工程学报, 34(4):299-305. |
| ZHAO JIE, YOU XINYONG, XU ZHENZHEN , et al, 2018. Review on application of SNP detection methods in animal research[J]. Transactions of the Chinese Society of Agricultural Engineering, 34(4):299-305 (in Chinese with English abstract). | |
| 32 | 赵琼一, 李信, 周德贵 , 等, 2010. 后基因组时代下作物的SNP分型方法[J]. 分子植物育种, 8(1):125-133. |
| ZHAO QIONGYI, LIN XIN, ZHOU DEGUI , et al, 2010. SNP genotyping methods for crops in post-genomic era[J]. Molecular Plant Breeding, 8(1):125-133 (in Chinese with English abstract). | |
| 33 | BOTSTEIN D, WHITE R L, SKOLNICK M , et al, 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms[J]. American Journal of Human Genetics, 32(3):314-331. |
| 34 |
GANAL M W, ALTMANN T, RÖDER M S , 2009. SNP identification in crop plants[J]. Current Opinion in Plant Biology, 12(2):211-217.
doi: 10.1016/j.pbi.2008.12.009 |
| 35 |
HINE P M, THORNE T , 2000. A survey of some parasites and diseases of several species of bivalve mollusc in northern Western Australia[J]. Diseases of Aquatic Organisms, 40(1):67-78.
doi: 10.3354/dao040067 |
| 36 |
HONG S P, SHIN S K, LEE E H , et al, 2008. High-resolution human papillomavirus genotyping by MALDI-TOF mass spectrometry[J]. Nature Protocols, 3(9):1476-1484.
doi: 10.1038/nprot.2008.136 |
| 37 |
LIEW M, PRYOR R, PALAIS R , et al, 2004. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons[J]. Clinical Chemistry, 50(7):1156-1164.
doi: 10.1373/clinchem.2004.032136 |
| 38 |
LIU WENGUANG, HUANG XIANDE, LIN JIANSHI , et al, 2012. Seawater acidification and elevated temperature affect gene expression patterns of the pearl oyster Pinctada fucata[J]. PLoS One, 7(3):e33679.
doi: 10.1371/journal.pone.0033679 |
| 39 |
MIYAKE T, ISOWA K, ISHIKAWA T , et al, 2016. Evaluation of genetic characteristics of wild and cultured populations of the Japanese pearl oyster Pinctada fucata martensii by using AFLP markers[J]. Aquaculture International, 24(2):537-548.
doi: 10.1007/s10499-015-9943-2 |
| 40 |
MIYAZAKI T, GOTO K, KOBAYASHI T , et al, 1999. Mass mortalities associated with a virus disease in Japanese pearl oysters Pinctada fucata martensii[J]. Diseases of Aquatic Organisms, 37(1):1-12.
doi: 10.3354/dao037001 |
| 41 |
MONTGOMERY J, WITTWER C T, PALAIS R , et al, 2007. Simultaneous mutation scanning and genotyping by high- resolution DNA melting analysis[J]. Nature Protocols, 2(1):59-66.
doi: 10.1038/nprot.2007.10 |
| 42 |
NEFF M M, NEFF J D, CHORY J , et al, 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics[J]. The Plant Journal, 14(3):387-392.
doi: 10.1046/j.1365-313X.1998.00124.x |
| 43 |
NEWTON C R, GRAHAM A, HEPTINSTALL L E , et al, 1989. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS)[J]. Nucleic Acids Research, 17(7):2503-2516.
doi: 10.1093/nar/17.7.2503 |
| 44 |
NICKERSON M L, WEIRICH G, ZBAR B , et al, 2000. Signature-based analysis of MET proto-oncogene mutations using DHPLC[J]. Human Mutation, 16(1):68-76.
doi: 10.1002/(ISSN)1098-1004 |
| 45 |
REED G H, WITTWER C T , 2004. Sensitivity and specificity of single-nucleotide polymorphism scanning by high- resolution melting analysis[J]. Clinical Chemistry, 50(10):1748-1754.
doi: 10.1373/clinchem.2003.029751 |
| 46 |
SHAO KE, XIONG MEIHUA, XU NIAN , et al, 2013. Characterization of microsatellite loci in Sinilabeo rendahli and cross-amplification in four other Chinese cyprinid species[J]. Conservation Genetics Resource, 5(1):9-13.
doi: 10.1007/s12686-012-9717-3 |
| 47 |
SOBRINO B, BRIÓN M, CARRACEDO A , 2005. SNPs in forensic genetics: a review on SNP typing methodologies[J]. Forensic Science International, 154(2-3):181-194.
doi: 10.1016/j.forsciint.2004.10.020 |
| 48 |
TAMURA K, PETERSON D, PETERSON N , et al, 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods[J]. Molecular Biology and Evolution, 28(10):2731-2739.
doi: 10.1093/molbev/msr121 |
| 49 | WADA K T, KOMARU A , 1991. Estimation of genetic variation in shell traits of the Japanese pearl oyster[J]. Bulletin of National Research Institute of Aquaculture.( Japan)/Yoshokukenho, ( 20):19-24. |
| 50 |
WADA K T, KOMARU A , 1994. Effect of selection for shell coloration on growth rate and mortality in the Japanese pearl oyster, Pinctada fucata martensii[J]. Aquaculture, 125(1-2):59-65.
doi: 10.1016/0044-8486(94)90282-8 |
| 51 | WEIR B S, COCKERHAM C C , 1984. Estimating F-statistics for the analysis of population structure[J]. Evolution, 38(6):1358-1370. |
| 52 |
WITTWER C T , 2009. High-resolution DNA melting analysis: Advancements and limitations[J]. Human Mutation, 30(6):857-859.
doi: 10.1002/humu.v30:6 |
| 53 | WRIGHT S , 1931. Evolution in Mendelian populations[J]. Genetics, 16(2):97-159. |
| 54 | WRIGHT S , 1978. Evolution and the genetics of populations, vol. 4. variability within and among Populations[M]. Chicago: University of Chicago Press. |
| 55 | YEH F C, YANG R C, BOYLE T , et al, 1999. POPGENE, version 1.32: The user friendly software for population genetic analysis[CP]. Canada: University of Alberta. |
| 56 | YOKOGAWA K , 1998. Morphological variabilities and genetic features in Japanese common clam Ruditapes philippinarum[J]. Venus (Japanese Journal of Malacology), 57(2):121-132. |
| 57 |
YU DAHUI, CHU KAHOU , 2006. Low genetic differentiation among widely separated populations of the pearl oyster Pinctada fucata as revealed by AFLP[J]. Journal of Experimental Marine Biology and Ecology, 333(1):140-146.
doi: 10.1016/j.jembe.2005.12.046 |
| [1] | LI Nenghui, HUANG Qing, LI Hang, ZENG Jun, WU Kefeng, TAN Huaqiang. Taxonomic study of four species of Gracilaria (Gracilariaceae, Rhodophyta) in Zhanjiang based on morphological and molecular data [J]. Journal of Tropical Oceanography, 2024, 43(2): 34-47. |
| [2] | LI Yunpeng, WANG Zhicheng, ZHANG Long, ZHANG Wei, DU Mengda, SUN Yi, ZHANG Zhifeng. Development of gonads with annual cycle in Anlactinia sinensis [J]. Journal of Tropical Oceanography, 2023, 42(6): 120-126. |
| [3] | ZHENG Guobin, ZHAO Hongbo, HUANG Liangmin, ZHANG Jing, LIU Xiande. Discovery and verification of SNP in Acanthopagrus latus [J]. Journal of Tropical Oceanography, 2023, 42(2): 78-86. |
| [4] | HAN Tong, LI Jingjing, LIU Zhengyi, LIU Kai, Zhang Jinhao, QIN Song, ZHONG Zhihai. The seed morphology and internal characteristics of seagrass, surfgrass Phyllospadix iwatensis [J]. Journal of Tropical Oceanography, 2022, 41(6): 105-113. |
| [5] | XU Hanzhi, ZHANG Hua, XIONG Panpan, HE Maoxian. Differential responses of allometric individuals to immune stimuli in Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2022, 41(5): 180-188. |
| [6] | GUO Junli, SHI Lianqiang, CHEN Shenliang, ZHANG Min, CHANG Yang, ZHANG Daheng. Dynamic variations of different sedimentary geomorphology of sandy and gravel embayed beaches on the Zhujiajian Island during typhoon season [J]. Journal of Tropical Oceanography, 2022, 41(4): 82-96. |
| [7] | LIU Jinmei, HUANG Bingxin, DING Lanping, WANG Xuecong, YAN Jing, YAN Panzhu, ZHANG Yao. Morphological and phylogenetic studies of Yonagunia formosana in southeastern Hainan province, China [J]. Journal of Tropical Oceanography, 2021, 40(6): 76-82. |
| [8] | LI Yonghang, JIA Lei, NI Yugen, HE Jian, MU Zelin, WEN Mingming, SHAN Chenchen. Geophysical characteristics and favorable occurrence signs of marine sand-gravel body in Taiwan banks* [J]. Journal of Tropical Oceanography, 2021, 40(5): 101-110. |
| [9] | WANG Ling, WANG Bin, LI Jian, YU Kaiqi, ZHAO Fang. Morphology characteristics and formation mechanisms of submarine pockmarks in the northern Zhongjiannan Basin, South China Sea [J]. Journal of Tropical Oceanography, 2021, 40(5): 72-84. |
| [10] | ZOU Congcong, WANG Lijuan, WU Zhihao, YOU Feng. Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences* [J]. Journal of Tropical Oceanography, 2021, 40(5): 25-35. |
| [11] | REN Huimin, ZHANG Heng, XU Shasha, LI Shengfa, LI Jiansheng, LI Zhihong, HE Lijun. Population genetic structure and historical dynamics of Trachurus japonicus in the China seas based on mitochondrial control region [J]. Journal of Tropical Oceanography, 2021, 40(5): 36-44. |
| [12] | NIU Biaobiao, ZHAI Mengyi, LI Yang. Morphological and Phylogenetic Studies on Chaetoceros seychellarus [J]. Journal of Tropical Oceanography, 2021, 40(4): 44-49. |
| [13] | LONG Chao, LUO Zhaohe, WEI Zhangliang, YANG Fangfang, LI Ru, LONG Lijuan. Morphology and phylogeny of zooxanthellae Effrenium voratum from Luhuitou reef in Sanya, Hainan province [J]. Journal of Tropical Oceanography, 2021, 40(4): 35-43. |
| [14] | DAI Zhijun, ZHOU Xiaoyan, WANG Jie, HU Baoqing. Review and prospect of mangrove tidal flat sedimentary dynamics [J]. Journal of Tropical Oceanography, 2021, 40(3): 69-75. |
| [15] | PAN Xiaolan, LIU Huiru, XU Meng, XU Hanzhi, ZHANG Hua, HE Maoxian. Preliminary study on immunity function of aquaporin AQP4 in the pearl oyster Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2021, 40(2): 83-89. |
|
||