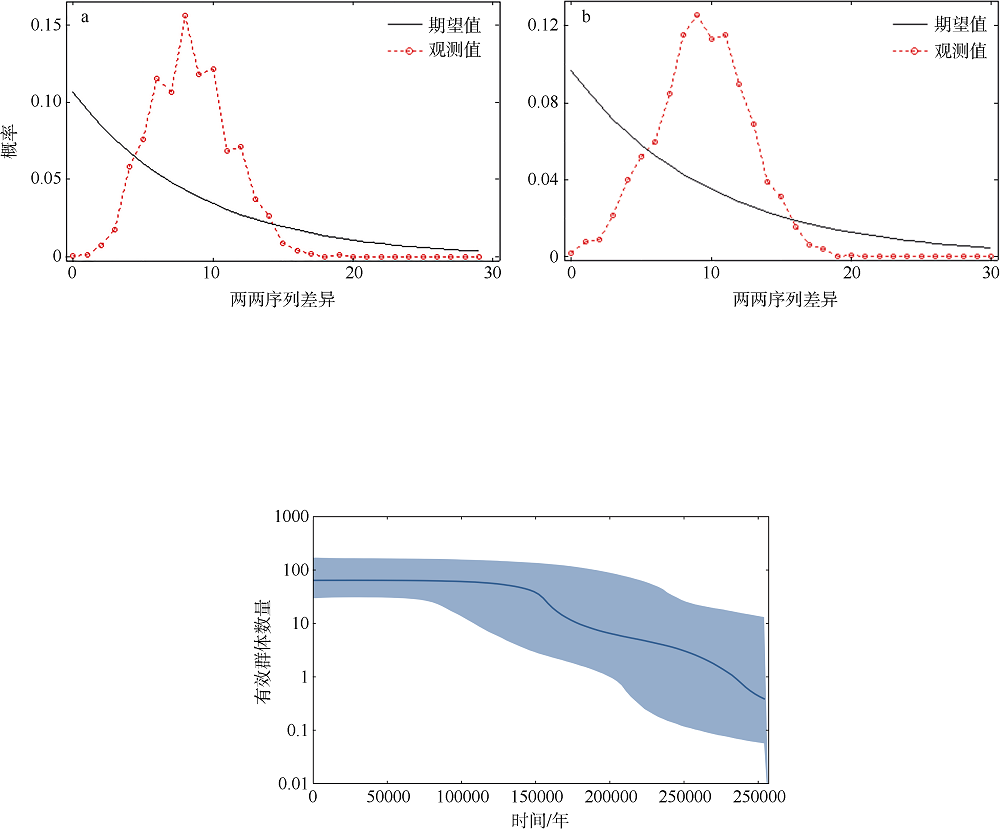
Journal of Tropical Oceanography ›› 2021, Vol. 40 ›› Issue (5): 25-35.doi: 10.11978/2020119CSTR: 32234.14.2020119
• Marine Biology • Previous Articles Next Articles
Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences*
ZOU Congcong1,2,3( ), WANG Lijuan1,2, WU Zhihao1,2, YOU Feng1,2(
), WANG Lijuan1,2, WU Zhihao1,2, YOU Feng1,2( )
)
- 1. Key Laboratory of Experimental Marine Biology, Center for Ocean Mega-Science, Institute of Oceanology, Chinese Academy of Sciences, Qingdao 266071, China
2. Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266237, China
3. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2020-10-15Revised:2021-01-29Online:2021-09-10Published:2021-02-09 -
Contact:YOU Feng E-mail:zoucongcong17@mails.ucas.ac.cn;youfeng@qdio.ac.cn -
Supported by:Special Program for Basic Research of the Ministry of Science and Technology of China(2014FY110500);National Marine Genetic Resource Center(2017DKA30470)
CLC Number:
- P735.54
Cite this article
ZOU Congcong, WANG Lijuan, WU Zhihao, YOU Feng. Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences*[J].Journal of Tropical Oceanography, 2021, 40(5): 25-35.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
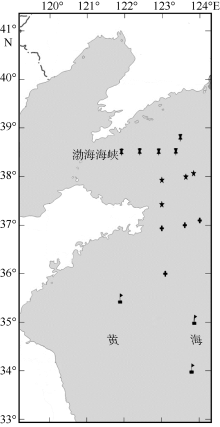
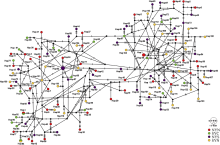
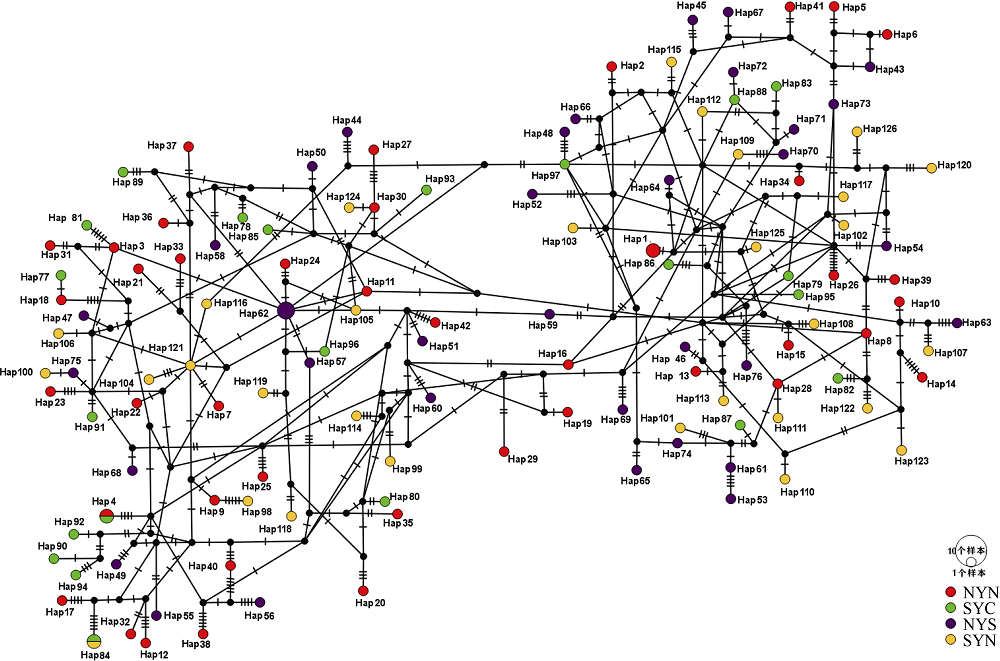
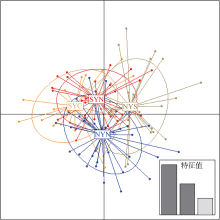
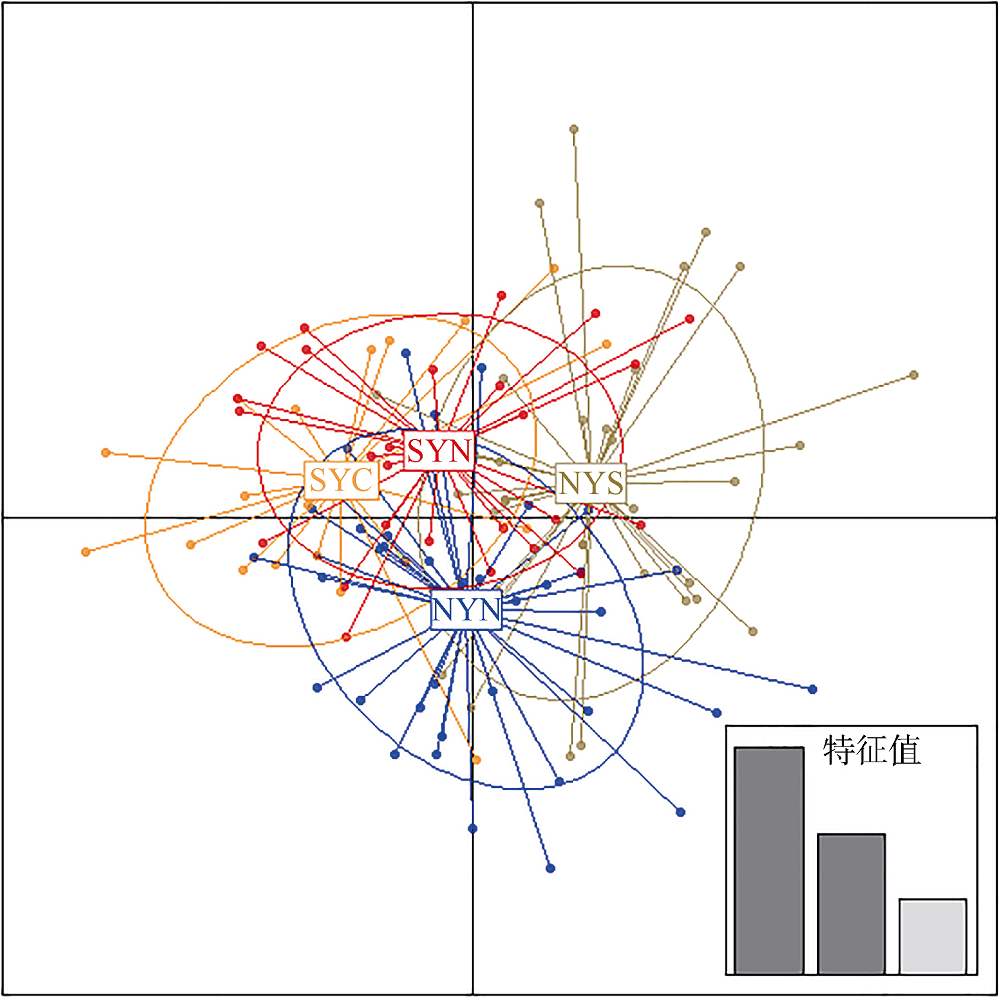
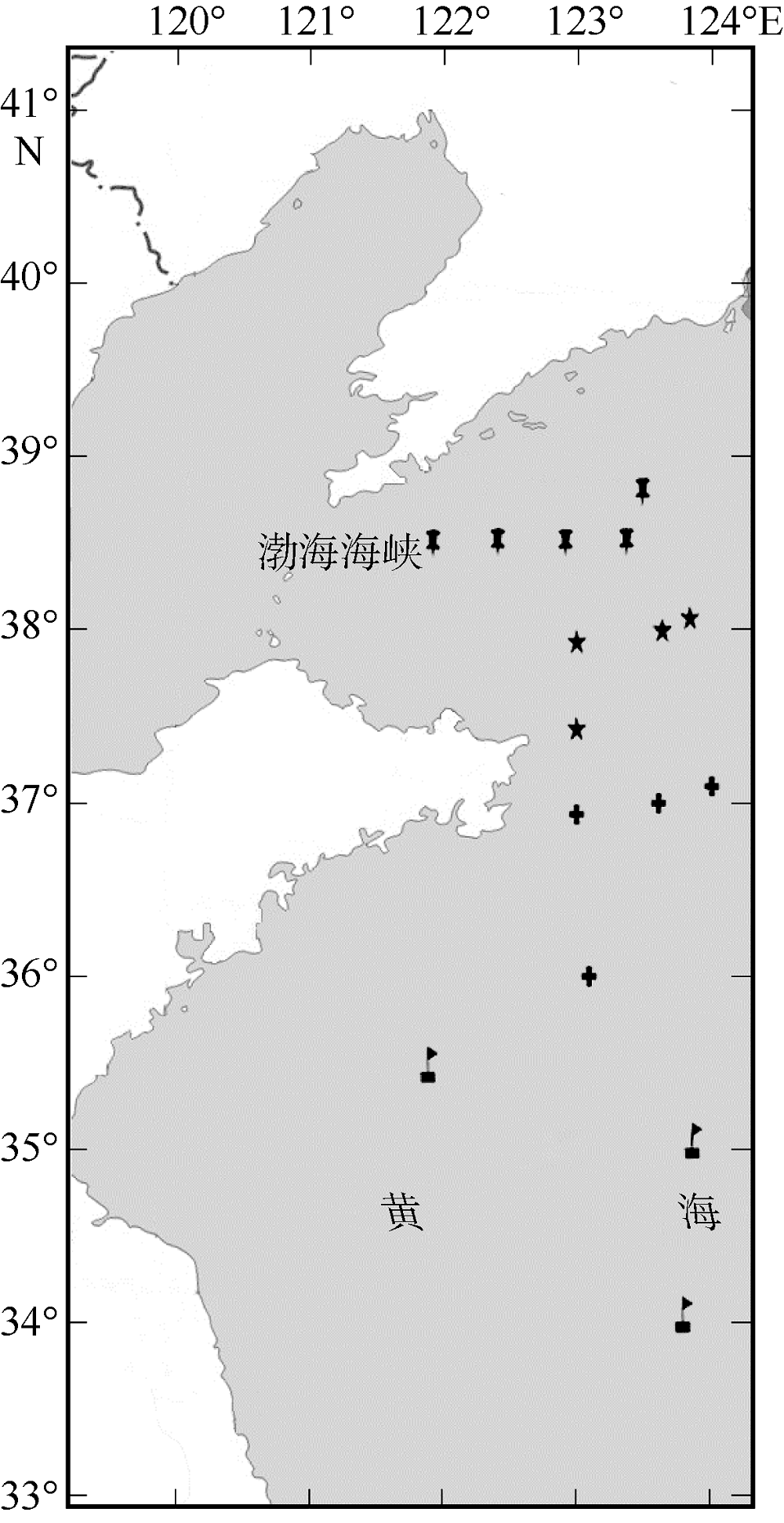
Fig. 1
Map of sampling locations of Engraulis japonicus in the Yellow Sea. , north of the Northern Yellow Sea population (NYN); , south of the Northern Yellow Sea population (NYS); , north of the Southern Yellow Sea population (SYN); , central part of the Southern Yellow Sea population (SYC)"
Tab. 1
Genetic diversity parameters in four Engraulis japonicus populations"
| 群体 | 样品数/个 | 单倍型数/个 | 单倍型多样性 (h±SD) | 核苷酸多样性 (π±SD) | 多态位点数/个 | 碱基转换数/个 | 碱基颠换数/个 | 碱基插入数/个 |
|---|---|---|---|---|---|---|---|---|
| NYN | 43 | 42 | 0.999±0.005 | 0.011±0.005 | 71 | 57 | 19 | 0 |
| NYS | 36 | 34 | 0.995±0.009 | 0.010±0.005 | 61 | 48 | 15 | 4 |
| SYN | 30 | 30 | 1.000±0.009 | 0.010±0.005 | 60 | 52 | 15 | 0 |
| SYC | 22 | 22 | 1.000±0.014 | 0.011±0.006 | 50 | 39 | 11 | 1 |
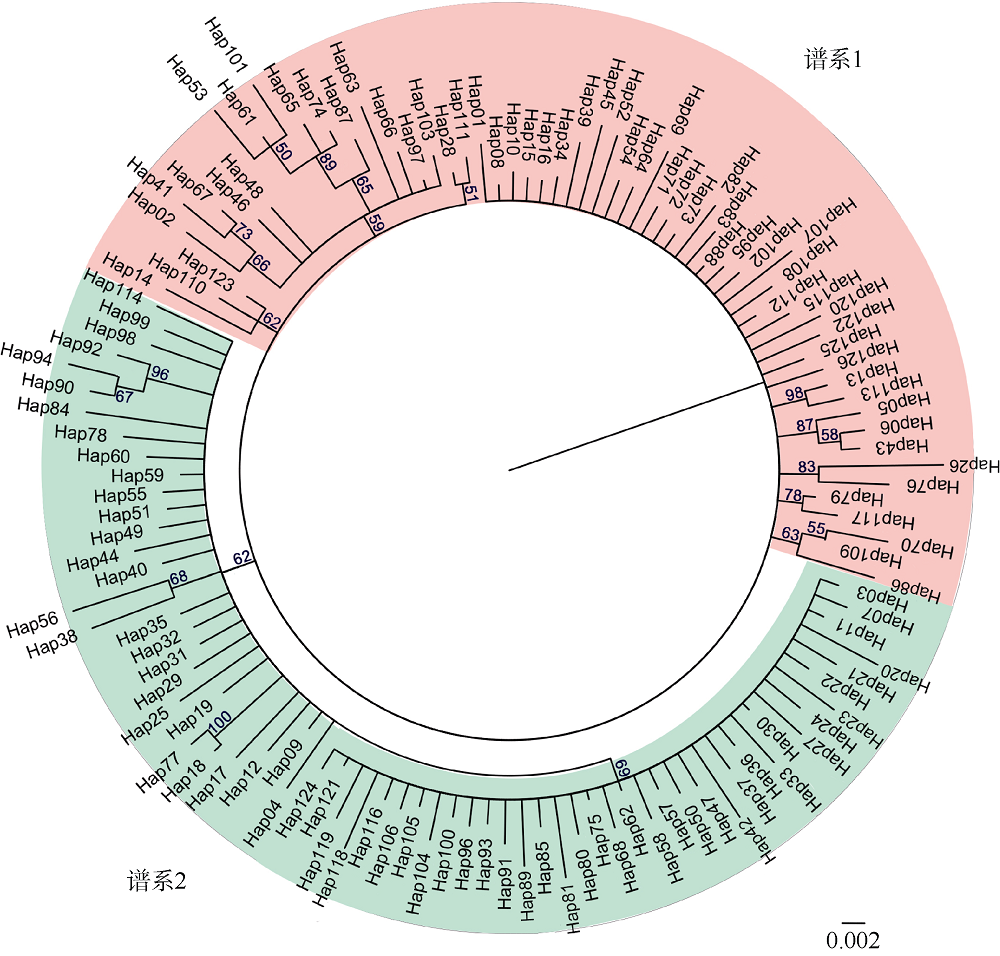
| 谱系1 | 61 | 60 | 1.000±0.003 | 0.008±0.004 | 72 | 59 | 23 | 1 |
| 谱系2 | 70 | 66 | 0.998±0.003 | 0.009±0.005 | 81 | 65 | 21 | 3 |
| 所有个体 | 131 | 126 | 0.999±0.001 | 0.010±0.005 | 108 | 85 | 38 | 4 |
Tab. 3
Analysis of Molecular Variance (AMOVA) in hierarchical structure"
| 变异来源 | 平方和 | 变异百分比 | F 统计 | p |
|---|---|---|---|---|
| 一个基因池 | ||||
| 群体间 | 16.901 | 0.190 | ||
| 群体内 | 673.710 | 99.810 | Fst = 0.00192 | 0.341 |
| 两个基因池(NYN、NYS)(SYN、SYC) | ||||
| 组群间 | 3.759 | -0.84 | Fct = -0.00841 | 1.000 |
| 组群内群体间 | 13.141 | 0.74 | Fsc = 0.00734 | 0.131 |
| 群体内 | 673.710 | 100.10 | Fst = -0.00101 | 0.286 |
| 两个基因池(NYN、NYS、SYN)(SYC) | ||||
| 组群间 | 6.157 | 0.41 | Fct = 0.00414 | 0.483 |
| 组群内群体间 | 10.743 | 0.03 | Fsc = 0.00035 | 0.392 |
| 群体内 | 673.710 | 99.55 | Fst = 0.00448 | 0.329 |
Tab. 4
Neutrality tests of four Engraulis japonicus populations in the Yellow Sea"
| 群体 | Tajima’s D | Fu’s Fs | Ramos-Onnsis and Roza’s R2 | |||
|---|---|---|---|---|---|---|
| D | p | Fs | p | R2 | p | |
| NYN | -1.307 | 0.073 | -24.603 | 0.000 | 0.107 | 0.000 |
| NYS | -1.013 | 0.162 | -24.077 | 0.000 | 0.113 | 0.000 |
| SYN | -1.348 | 0.070 | -24.249 | 0.000 | 0.118 | 0.000 |
| SYC | -0.702 | 0.284 | -13.580 | 0.000 | 0.127 | 0.000 |
| 谱系 1 | -1.532 | 0.034 | -24.790 | 0.000 | 0.103 | 0.000 |
| 谱系 2 | -1.435 | 0.041 | -24.619 | 0.000 | 0.099 | 0.000 |
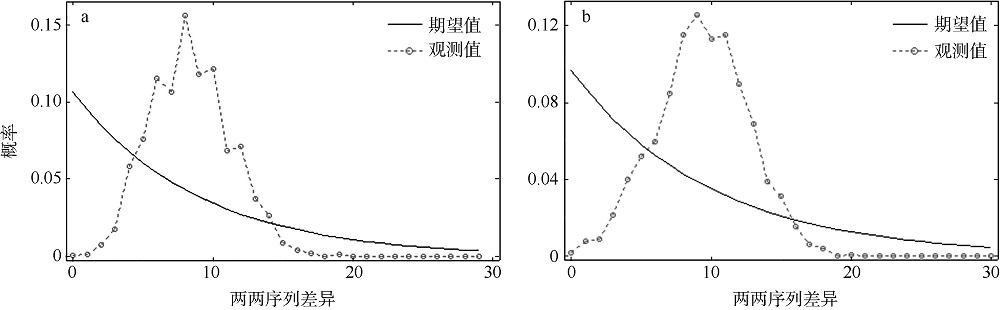
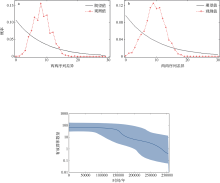
Tab. 5
Mismatch distribution under demographic expansion and spatial expansion models"
| 群体 | 群体扩张模型 | 空间扩张模型 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| τ | t | θ0 | θ1 | SSD | HRI | τ | θ | M | SSD | HRI | |
| NYN | 10.578 | 0.172 | 0.000 | 1527.500 | 0.001 | 0.007 | 10.585 | 0.010 | 1574.756 | 0.001 | 0.007 |
| NYS | 9.396 | 0.153 | 1.331 | 386.250 | 0.002 | 0.009 | 9.232 | 1.480 | 206.480 | 0.002 | 0.009 |
| SYN | 9.855 | 0.160 | 0.000 | 280.625 | 0.003 | 0.011 | 8.745 | 1.201 | 99999.000 | 0.003 | 0.011 |
| SYC | 11.881 | 0.193 | 0.002 | 279.688 | 0.004 | 0.010 | 11.802 | 0.026 | 99999.000 | 0.004 | 0.010 |
| 谱系1 | 8.326 | 0.135 | 0.228 | 99999.000 | 0.001 | 0.010 | 8.543 | 0.103 | 99999.000 | 0.001 | 0.010 |
| 谱系2 | 10.109 | 0.164 | 0.025 | 118.896 | 0.001 | 0.005 | 9.189 | 0.860 | 287.179 | 0.001 | 0.005 |
| [1] | 成庆泰, 郑葆珊, 1987. 中国鱼类系统检索[M]. 北京: 科学出版社: 59-61. |
| CHENG QINGTAI, ZHENG BAOSHAN, 1987. Systematic synopsis of Chinese fishes[M]. Beijing: Science Press: 59-61(in Chinese). | |
| [2] | 高天翔, 高兵兵, 李忠炉, 等, 2020. 基于线粒体DNA控制区序列的短棘鲾群体遗传学[J]. 水产学报, 44(5):715-722. |
| GAO TIANXIANG, GAO BINGBING, LI ZHONGLU, et al, 2020. Population genetics study of Leiognathus equulus based on the control region fragment of mitochondrial DNA[J]. Journal of Fisheries of China, 44(5):715-722 (in Chinese with English abstract). | |
| [3] | 胡东方, 2009. 黄海鳀鱼的摄食生态学研究[D]. 青岛: 中国海洋大学: 1-2. |
| HU DONGFANG, 2009. Studies on feeding ecology of anchovy in Yellow Sea[D]. Qingdao: Ocean University of China: 1-2 (in Chinese with English abstract). | |
| [4] | 李秀梅, 叶振江, 李增光, 等, 2016. 黄海中部近岸产卵场日本鳀卵子大小的时空变化[J]. 中国海洋大学学报, 46(2):54-60. |
| LI XIUMEI, YE ZHENJIANG, LI ZENGGUANG, et al, 2016. Spatial and temporal changes of the egg Size of Japanese Anchovy (Engraulis japonicus) inhabiting the near shore spawning ground of central Yellow Sea[J]. Periodical of Ocean University of China, 46(2):54-60 (in Chinese with English abstract). | |
| [5] | 梁述章, 宋炜, 马春艳, 等, 2019. 基于线粒体控制区的中国近海棘头梅童鱼群体遗传结构研究[J]. 海洋渔业, 41(22):138-148. |
| LIANG SHUZHANG, SONG WEI, MA CHUNYAN, et al, 2019. Genetic structure of Collichthys lucidus populations from China coastal waters based on mt DNA control region[J]. Marine Fisheries, 41(2):138-148 (in Chinese with English abstract). | |
| [6] | 唐启升, 苏纪兰, 2000. 中国海洋生态系统动力学研究. Ⅰ. 关键科学问题与研究发展战略[M]. 北京: 科学出版社: 45-72(in Chinese). |
| [7] | 吴仁协, 张浩冉, 牛素芳, 等, 2019. 东海近岸带鱼(Trichiurus japonicus)线粒体控制区序列的群体遗传变异研究[J]. 海洋与湖沼, 50(6):1318-1327. |
| WU RENXIE, ZHANG HAORAN, NIU SUFANG, et al, 2019. Study on population genetic variation of Trichiurus japonicus in nearshore of the East China Sea in mitochondrial control region sequences[J]. Oceanologia et Limnologia Sinica, 50(6):1318-1327 (in Chinese with English abstract). | |
| [8] | 薛利建, 刘子藩, 2005. 东海日本鳀数量分布和生物学特性的研究[J]. 浙江海洋学院学报(自然科学版), 24(4):312-317. |
| XUE LIJIAN, LIU ZIFAN, 2005. Study on the biomass distribution & biological characteristics of Engraulis japonicus in the East China Sea[J]. Journal of Zhejiang Ocean University (Natural Science), 24(4):312-317 (in Chinese with English abstract). | |
| [9] | 赵爽, 章群, 乐小亮, 等, 2010. 中国近海5个黑鲷地理群体的遗传变异[J]. 海洋科学, 34(2):75-79. |
| ZHAO SHUANG, ZHANG QUN, YUE XIAOLIANG, et al, 2010. Genetic variation among 5 stocks of Acanthopagrus schlegeli in China’s coastal waters[J]. Marine Sciences, 34(2):75-79 (in Chinese with English abstract). | |
| [10] | 赵宪勇, 2006. 黄海鳀鱼种群动力学特征及其资源可持续利用[D]. 青岛: 中国海洋大学: 18-43. |
| ZHAO XIANYONG, 2006. Population dynamic characteristics and sustainable utilization of the anchovy in the Yellow Sea[D]. Qingdao: Ocean University of China: 18-43 (in Chinese with English abstract). | |
| [11] | 朱琨, 刘微, 2020. 一种鳀鱼提取物的开发和生产工艺研究[J]. 中国食品添加剂, 31(5):69-74. |
| ZHU KUN, LIU WEI, 2020. The development and technology of anchovy extract production[J]. China Food Additives, 31(5):69-74 (in Chinese with English abstract). | |
| [12] |
BOUCKAERT R, HELED J, KÜHNERT D, et al, 2014. Beast 2: a software platform for bayesian evolutionary analysis[J]. PLoS Computational Biology, 10(4):e1003537.
doi: 10.1371/journal.pcbi.1003537 |
| [13] |
CANALES-AGUIRRE C B, FERRADA-FUENTES S, GALLEGUILLOS R, et al, 2018. High genetic diversity and low-population differentiation in the patagonian sprat (Sprattus fuegensis) based on mitochondrial DNA[J]. Mitochondrial DNA Part A, 29(8):1148-1155.
doi: 10.1080/24701394.2018.1424841 |
| [14] |
CHECKLEY JR D M, ASCH R G, RYKACZEWSKI R R, 2017. Climate, anchovy, and sardine[J]. Annual Review of Marine Science, 9:469-493.
doi: 10.1146/annurev-marine-122414-033819 |
| [15] |
CHENG JIAO, HAN ZHIQIANG, SONG NA, et al, 2018. Effects of Pleistocene glaciation on the phylogeographic and demographic histories of chub mackerel Scomber japonicus in the north-western Pacific[J]. Marine and Freshwater Research, 69(4):514-524.
doi: 10.1071/MF17099 |
| [16] |
CLARKE C R, KARL S A, HORN R L, et al, 2015. Global mitochondrial DNA phylogeography and population structure of the silky shark, Carcharhinus falciformis[J]. Marine Biology, 162(5):945-955.
doi: 10.1007/s00227-015-2636-6 |
| [17] | DARRIBA D, TABOADA G L, DOALLO R, et al, 2012. jModelTest 2: more models, new heuristics and parallel computing[J]. Nature Methods, 9(8):772. |
| [18] |
DOMINGUES R R, HILSDORF A W S, SHIVJI M M, et al, 2018. Effects of the Pleistocene on the mitochondrial population genetic structure and demographic history of the silky shark (Carcharhinus falciformis) in the western Atlantic Ocean[J]. Reviews in Fish Biology and Fisheries, 28(1):213-227.
doi: 10.1007/s11160-017-9504-z |
| [19] |
DONALDSON K A, WILSON JR R R, 1999. Amphi-panamic geminates of snook (Percoidei: Centropomidae) provide a calibration of the divergence rate in the mitochondrial DNA control region of fishes[J]. Molecular Phylogenetics and Evolution, 13(1):208-213.
doi: 10.1006/mpev.1999.0625 |
| [20] |
EXCOFFIER L, LISCHER H E L, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows[J]. Molecular Ecology Resources, 10(3):564-567.
doi: 10.1111/men.2010.10.issue-3 |
| [21] |
GRANT W A S, BOWEN B W, 1998. Shallow population histories in deep evolutionary lineages of marine fishes: Insights from sardines and anchovies and lessons for conservation[J]. Journal of Heredity, 89(5):415-426.
doi: 10.1093/jhered/89.5.415 |
| [22] |
GUO SHANSHAN, ZHANG GUIRONG, GUO XIANGZHAO, et al, 2014. Genetic diversity and population structure of Schizopygopsis younghusbandi Regan in the Yarlung Tsangpo River inferred from mitochondrial DNA sequence analysis[J]. Biochemical Systematics and Ecology, 57:141-151.
doi: 10.1016/j.bse.2014.07.026 |
| [23] |
HAN ZHIQIANG, GAO TIANXIANG, YANAGIMOTO T, et al, 2008. Genetic population structure of Nibea albiflora in Yellow Sea and East China Sea[J]. Fisheries Science, 74(3):544-552.
doi: 10.1111/j.1444-2906.2008.01557.x |
| [24] |
HAYASHI A, ZHANG K, SARUWATARI T, et al, 2016. Distribution of eggs and larvae of Japanese anchovy Engraulis japonicus in the Pacific waters off northern Japan in summer[J]. Fisheries Science, 82(2):311-319.
doi: 10.1007/s12562-015-0966-4 |
| [25] |
INOUE K, MONROE E M, ELDERKIN C L, et al, 2014. Phylogeographic and population genetic analyses reveal Pleistocene isolation followed by high gene flow in a wide ranging, but endangered, freshwater mussel[J]. Heredity, 112(3):282-290.
doi: 10.1038/hdy.2013.104 |
| [26] |
JAMALUDIN N A, MOHD-ARSHAAD W, AKIB N A M, et al, 2020. Phylogeography of the Japanese scad, Decapterus maruadsi (Teleostei; Carangidae) across the Central Indo-West Pacific: evidence of strong regional structure and cryptic diversity[J]. Mitochondrial DNA Part A, 31(7):298-310.
doi: 10.1080/24701394.2020.1799996 |
| [27] |
JUNG S, PANG I-C, LEE J-H, et al, 2016. Climate-change driven range shifts of anchovy biomass projected by bio-physical coupling individual based model in the marginal seas of East Asia[J]. Ocean Science Journal, 51(4):563-580.
doi: 10.1007/s12601-016-0055-3 |
| [28] |
KASIM N S, JAAFAR T N A M, PIAH R M, et al, 2020. Recent population expansion of longtail tuna Thunnus tonggol (Bleeker, 1851) inferred from the mitochondrial DNA markers[J]. PeerJ, 8:e9679.
doi: 10.7717/peerj.9679 |
| [29] |
KETCHUM R N, DIENG M M, VAUGHAN G O, et al, 2016. Levels of genetic diversity and taxonomic status of Epinephelus species in United Arab Emirates fish markets[J]. Marine Pollution Bulletin, 105(2):540-545.
doi: 10.1016/j.marpolbul.2015.11.042 |
| [30] |
KIM J Y, CHO E S, KIM W J, 2004. Population genetic structure of Japanese Anchovy (Engraulis japonicus) in Korean waters based on mitochondrial 12S ribosomal RNA gene sequences[J]. Journal of Life Science, 14(6):938-950.
doi: 10.5352/JLS.2004.14.6.938 |
| [31] |
KWON D-H, HWANG S-D, LIM D, 2012. Catch predictions for Pacific anchovy Engraulis japonicus larvae in the Yellow Sea[J]. Fisheries and Aquatic Sciences, 15(4):345-352.
doi: 10.5657/FAS.2012.0345 |
| [32] |
LARKIN M A, BLACKSHIELDS G, BROWN N P, et al, 2007. Clustal W and Clustal X version 2.0[J]. Bioinformatics, 23(21):2947-2948.
doi: 10.1093/bioinformatics/btm404 |
| [33] |
LEIGH J W, BRYANT D, 2015. POPART: full-feature software for haplotype network construction[J]. Methods in Ecology and Evolution, 6(9):1110-1116.
doi: 10.1111/mee3.2015.6.issue-9 |
| [34] |
LIU JINXIAN, GAO TIANXIANG, ZHUANG ZHIMENG, et al, 2006. Late Pleistocene divergence and subsequent population expansion of two closely related fish species, Japanese anchovy (Engraulis japonicus) and Australian anchovy (Engraulis australis)[J]. Molecular Phylogenetics and Evolution, 40(3):712-723.
doi: 10.1016/j.ympev.2006.04.019 |
| [35] |
LIU JINXIAN, GAO TIANXIANG, WU SHIFANG, et al, 2007. Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845)[J]. Molecular Ecology, 16(2):275-288.
doi: 10.1111/j.1365-294X.2006.03140.x |
| [36] |
MA CHUNYAN, CHENG QIQUN, ZHANG QINGYI, et al, 2010. Genetic variation of Coilia ectenes (Clupeiformes: Engraulidae) revealed by the complete cytochrome b sequences of mitochondrial DNA[J]. Journal of Experimental Marine Biology and Ecology, 385(1-2):14-19.
doi: 10.1016/j.jembe.2010.01.015 |
| [37] |
MENEZES M R, KUMAR G, KUNAL S P, 2012. Population genetic structure of skipjack tuna Katsuwonus pelamis from the Indian coast using sequence analysis of the mitochondrial DNA D-loop region[J]. Journal of Fish Biology, 80(6):2198-2212.
doi: 10.1111/j.1095-8649.2012.03270.x |
| [38] |
MONTES I, IRIONDO M, MANZANO C, et al, 2017. Development of gene-associated single nucleotide polymorphisms for Japanese anchovy Engraulis japonicus through cross-species amplification[J]. Fisheries Science, 84(1):1-7.
doi: 10.1007/s12562-017-1134-9 |
| [39] |
MUNIAN K, BHASSU S, 2015. Genetic structure of locally threatened cyprinid, Osteochilus melanopleurus, in Peninsular Malaysia River systems inferred from mitochondrial DNA control region[J]. Biochemical Systematics and Ecology, 61:336-343.
doi: 10.1016/j.bse.2015.06.034 |
| [40] | MYOUNG S H, KIM J-K, 2014. Genetic diversity and population structure of the gizzard shad, Konosirus punctatus (Clupeidae, Pisces), in Korean waters based on mitochondrial DNA control region sequences[J]. Genes & Genomics, 36(5):591-598. |
| [41] | NEI M, 1987. Molecular evolutionary genetics[M]. New York: Columbia University Press. |
| [42] |
NYKÄNEN M, DILLANE E, REID D, et al, 2020. Genetic methods reveal high diversity and no evidence of stock structure among cuckoo rays (Leucoraja naevus) in the northern part of Northeast Atlantic[J]. Fisheries Research, 232:105715.
doi: 10.1016/j.fishres.2020.105715 |
| [43] |
OHATA R, MASUDA R, YAMASHITA Y, 2018. Ontogeny of turbiditaxis in hatchery-reared Japanese anchovy Engraulis japonicus[J]. Journal of Fish Biology, 92(4):1235-1240.
doi: 10.1111/jfb.2018.92.issue-4 |
| [44] |
OTWOMA L M, REUTER H, TIMM J, et al, 2018. Genetic connectivity in a herbivorous coral reef fish (Acanthurus leucosternon Bennet, 1833) in the eastern African region[J]. Hydrobiologia, 806(1):237-250.
doi: 10.1007/s10750-017-3363-4 |
| [45] | PANDEY D, RYU Y-W, MATSUBARA T, 2017. Features of sperm motility and circadian rhythm in Japanese anchovy (Engraulis japonicus)[J]. Fisheries and Aquaculture Journal, 8(2):203. |
| [46] |
PEDROSA-GERASMIO I R, AGMATA A B, SANTOS M D, 2015. Genetic diversity, population genetic structure, and demographic history of Auxis thazard (Perciformes), Selar crumenophthalmus (Perciformes), Rastrelliger kanagurta (Perciformes) and Sardinella lemuru (Clupeiformes) in Sulu-Celebes Sea inferred by mitochondrial DNA sequences[J]. Fisheries Research, 162:64-74.
doi: 10.1016/j.fishres.2014.10.006 |
| [47] |
PILLANS B, CHAPPELL J, NAISH T R, 1998. A review of the Milankovitch climatic beat: template for Plio-Pleistocene sea-level changes and sequence stratigraphy[J]. Sedimentary Geology, 122(1-4):5-21.
doi: 10.1016/S0037-0738(98)00095-5 |
| [48] |
RONQUIST F, TESLENKO M, VAN DER MARK P, et al, 2012. MrBayes 3.2: efficient bayesian phylogenetic inference and model choice across a large model space[J]. Systematic Biology, 61(3):539-542.
doi: 10.1093/sysbio/sys029 |
| [49] |
SEFC K M, WAGNER M, ZANGL L, et al, 2020. Phylogeographic structure and population connectivity of a small benthic fish (Tripterygion tripteronotum) in the Adriatic Sea[J]. Journal of Biogeography, 47(11):2502-2517.
doi: 10.1111/jbi.v47.11 |
| [50] |
SHEN K-N, JAMANDRE B W, HSU C-C, et al, 2011. Plio-Pleistocene sea level and temperature fluctuations in the Northwestern Pacific promoted speciation in the globally-distributed flathead mullet Mugil cephalus[J]. BMC Evolutionary Biology, 11(1):83.
doi: 10.1186/1471-2148-11-83 |
| [51] |
SUN CHAO, XUAN ZHONGYA, LIU HONGBO, et al, 2019. Cyt b gene and D-loop sequence analyses of Coilia nasus from the Rokkaku River of Japan[J]. Regional Studies in Marine Science, 32:100840.
doi: 10.1016/j.rsma.2019.100840 |
| [52] | TAKAHASHI M, WATANABE Y, 2005. Effects of temperature and food availability on growth rate during late larval stage of Japanese anchovy (Engraulis japonicus) in the Kuroshio-Oyashio transition region[J]. Fisheries Oceanography, 14(3):223-235. |
| [53] |
WANG QIAN, ZHANG JIE, MATSUMOTO H, et al, 2016. Population structure of elongate ilisha Ilisha elongata along the Northwestern Pacific Coast revealed by mitochondrial control region sequences[J]. Fisheries Science, 82(5):771-785.
doi: 10.1007/s12562-016-1018-4 |
| [54] |
XING QINWANG, YU HUAMING, YU HAIQING, et al, 2020. A comprehensive model-based index for identification of larval retention areas: a case study for Japanese anchovy Engraulis japonicus in the Yellow Sea[J]. Ecological Indicators, 116:106479.
doi: 10.1016/j.ecolind.2020.106479 |
| [55] |
YU HAIQING, YU HUAMING, ITO S-I, et al, 2020. Potential environmental drivers of Japanese anchovy (Engraulis japonicus) recruitment in the Yellow Sea[J]. Journal of Marine Systems, 212:103431.
doi: 10.1016/j.jmarsys.2020.103431 |
| [56] |
YU ZINIU, KONG XIAOYU, GUO TIANHUI, et al, 2005. Mitochondrial DNA sequence variation of Japanese anchovy Engraulis japonicus from the Yellow Sea and East China Sea[J]. Fisheries Science, 71(2):299-307.
doi: 10.1111/fis.2005.71.issue-2 |
| [57] |
ZHANG YANHONG, PHAM N K, ZHANG HUIXIAN, et al, 2014. Genetic variations in two seahorse species (Hippocampus mohnikei and Hippocampus trimaculatus): Evidence for middle pleistocene population expansion[J]. PLoS One, 9(8):e105494.
doi: 10.1371/journal.pone.0105494 |
| [58] |
ZHANG YURONG, YANG FANG, WANG ZHAOLIN, et al, 2017. Mitochondrial DNA variation and population genetic structure in the small yellow croaker at the coast of Yellow Sea and East China Sea[J]. Biochemical Systematics and Ecology, 71:236-243.
doi: 10.1016/j.bse.2017.03.003 |
| [59] |
ZHENG WEI, ZOU LI, HAN ZHIQIANG, 2015. Genetic analysis of the populations of Japanese anchovy Engraulis japonicus from the Yellow Sea and East China Sea based on mitochondrial cytochrome b sequence[J]. Biochemical Systematics and Ecology, 58:169-177.
doi: 10.1016/j.bse.2014.12.007 |
| [60] |
ZHOU XIN, SUN YAO, HUANG WEN, et al, 2015. The Pacific decadal oscillation and changes in anchovy populations in the Northwest Pacific[J]. Journal of Asian Earth Sciences, 114:504-511.
doi: 10.1016/j.jseaes.2015.06.027 |
| [1] | REN Huimin, ZHANG Heng, XU Shasha, LI Shengfa, LI Jiansheng, LI Zhihong, HE Lijun. Population genetic structure and historical dynamics of Trachurus japonicus in the China seas based on mitochondrial control region [J]. Journal of Tropical Oceanography, 2021, 40(5): 36-44. |
| [2] | Min LI, Xiaolan KONG, Youwei XU, Zuozhi CHEN. Genetic polymorphism of the Brushtooth lizardfish Saurida undosquamis based on mitochondrial D-loop sequences [J]. Journal of Tropical Oceanography, 2020, 39(4): 42-49. |
| [3] | Xiaolin HUANG, Wenjun LI, Heizhao LIN, Yukai YANG, Tao LI, Wei YU, Zhong HUANG. Genetic variations among Siganus oramin populations in coastal waters of southeast China based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2018, 37(4): 45-51. |
| [4] | Zhiying ZHAO, Liyun LIANG, Lirong BAI. Analysis of genetic diversity among three wild populations of Penaeus monodon using microsatellite marker [J]. Journal of Tropical Oceanography, 2018, 37(3): 65-72. |
| [5] | Xiaoqing YIN, Dingtian YANG, Lizhu ZHOU. Estimation of suspended particulate matter transport via the boundary waters of the Yellow Sea and the East Sea based on satellite remote sensing [J]. Journal of Tropical Oceanography, 2018, 37(2): 10-16. |
| [6] | WENG Zhaohong, XIE Yangjie, XIAO Zhiqun, WANG Yilei. Analysis of genetic diversity in several wild and hatchery populations of Crassostrea angulata from south Fujian and south Guangdong [J]. Journal of Tropical Oceanography, 2016, 35(3): 94-98. |
| [7] | QI Jiang-hao, ZHANG Xun-hua, WU Zhi-qiang, QIU Xue-lin, ZHAO Ming-hui, XIA Shao-hong, GUO Xing-wei, HAO Tian-yao, ZHENG Yan-peng, FANG Nian-qiao. Preliminary results of the South Yellow Sea OBS 2013 onshore-offshore joint deep seismic survey [J]. Journal of Tropical Oceanography, 2015, 34(2): 76-84. |
| [8] | SHI Xiao-feng1, 2, SU Yong-quan1, WANG Wen-cheng1, WANG Jun1. Population genetic structure of three stocks of Acanthopagrus schlegelii based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2015, 34(1): 56-63. |
| [9] | HAN Xue, CAI Yi, CHEN Xing-rong, LI Yan. Analysis of temporal and spatial distribution characteristics of winter SST anomalies in the Bohai Sea and Yellow Sea and their influencing factors [J]. Journal of Tropical Oceanography, 2014, 33(5): 1-12. |
| [10] | LIU Li, ZHAO Jie, GUO Yu-song, LIU Chu-wu. Study on the AFLP primer selection and genetic diversities of 4 Snappers [J]. Journal of Tropical Oceanography, 2012, 31(4): 112-116. |
| [11] | LI Li-hao,YU Da-hui,HUANG Gui-ju,DU Bo,FU Yun,TONG Xin,GUO Yi-hui,YE Wei. Comparison of genetic diversity among stocks of Oreochromis niloticus, O. aureus and red tilapia based on microsatellite DNA [J]. Journal of Tropical Oceanography, 2012, 31(2): 102-109. |
| [12] | JI Lei,OU You-jun,LI Jia-er. Genetic polymorphism of three cultured populations of golden pompano Trachinotus ovatus as revealed by microsatellites [J]. Journal of Tropical Oceanography, 2011, 30(3): 62-68. |
| [13] | GUO Yu-song,WANG Zhong-duo,LIU Chu-wu,CHEN Zhi-ming,LIU Yun. Isolation and genetic diversity analysis of microsatellites from nine species of familiar snappers [J]. Journal of Tropical Oceanography, 2010, 29(3): 82-86. |
|
||