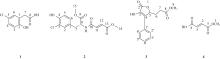
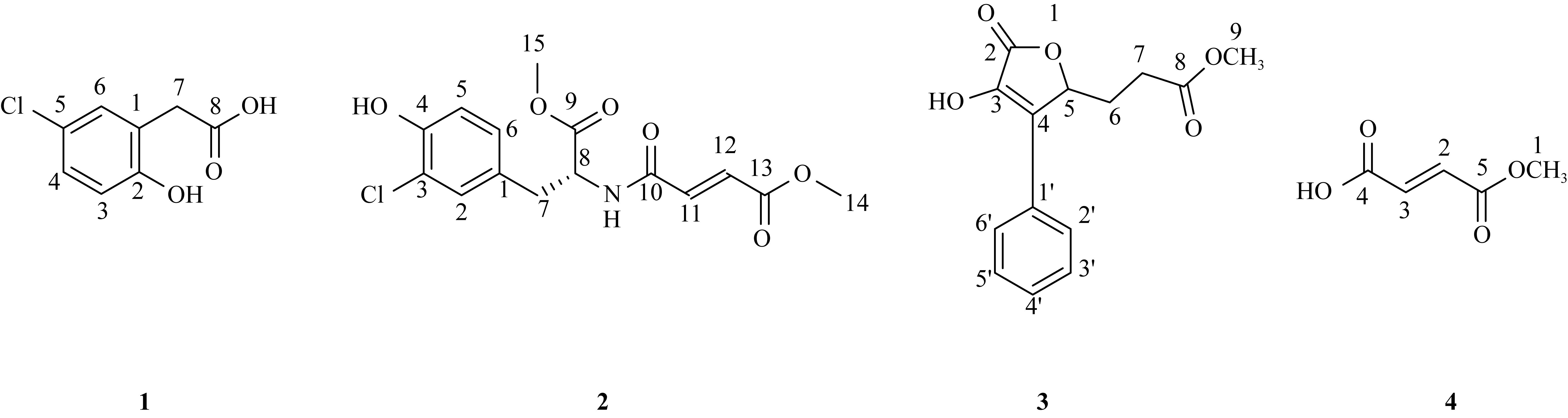
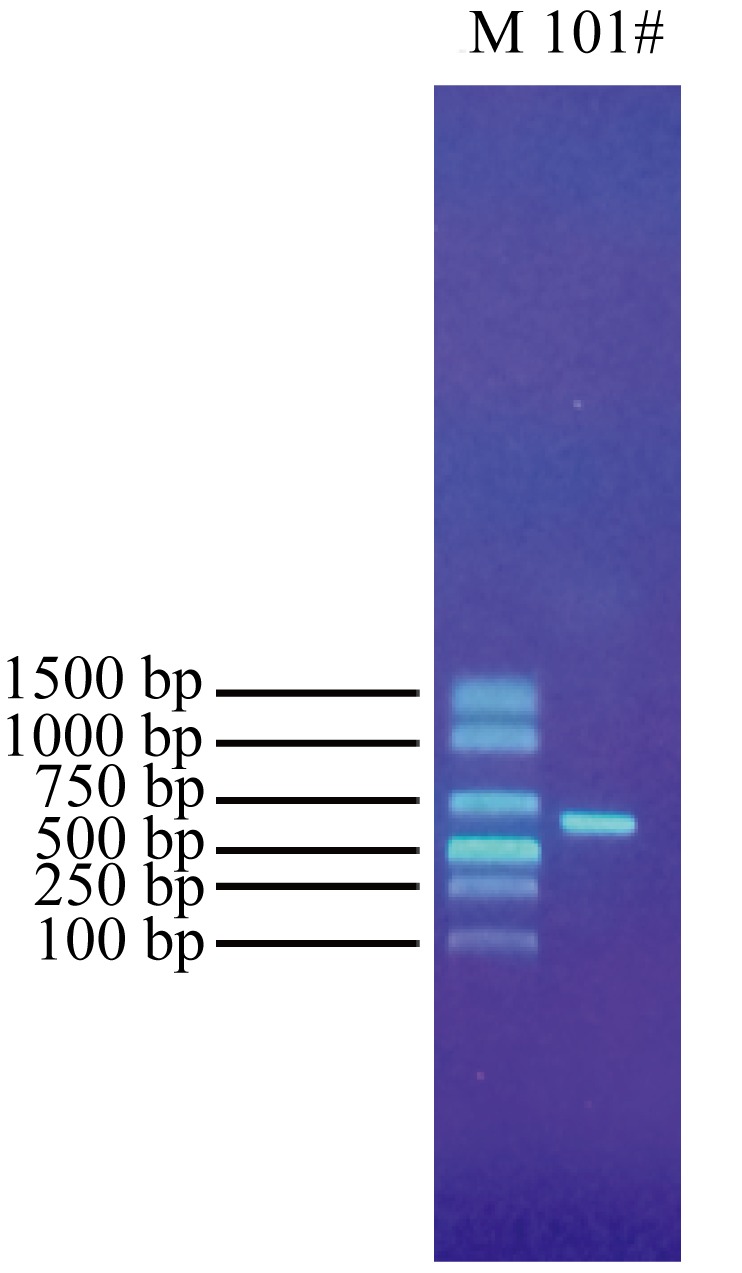
Journal of Tropical Oceanography ›› 2023, Vol. 42 ›› Issue (5): 104-114.doi: 10.11978/2022242CSTR: 32234.14.2022242
• Marine Biology • Previous Articles Next Articles
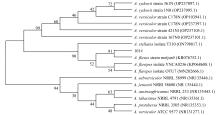
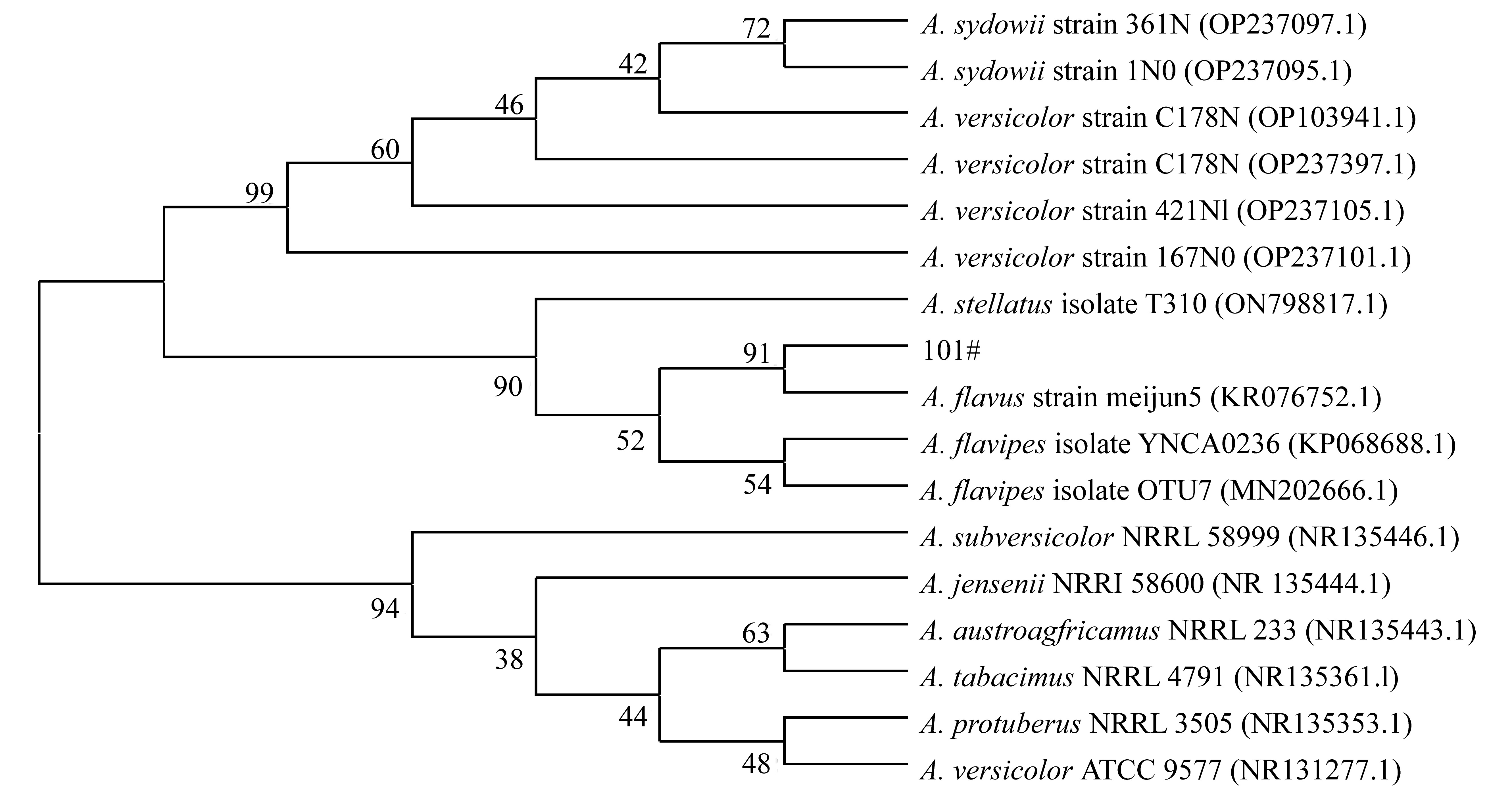
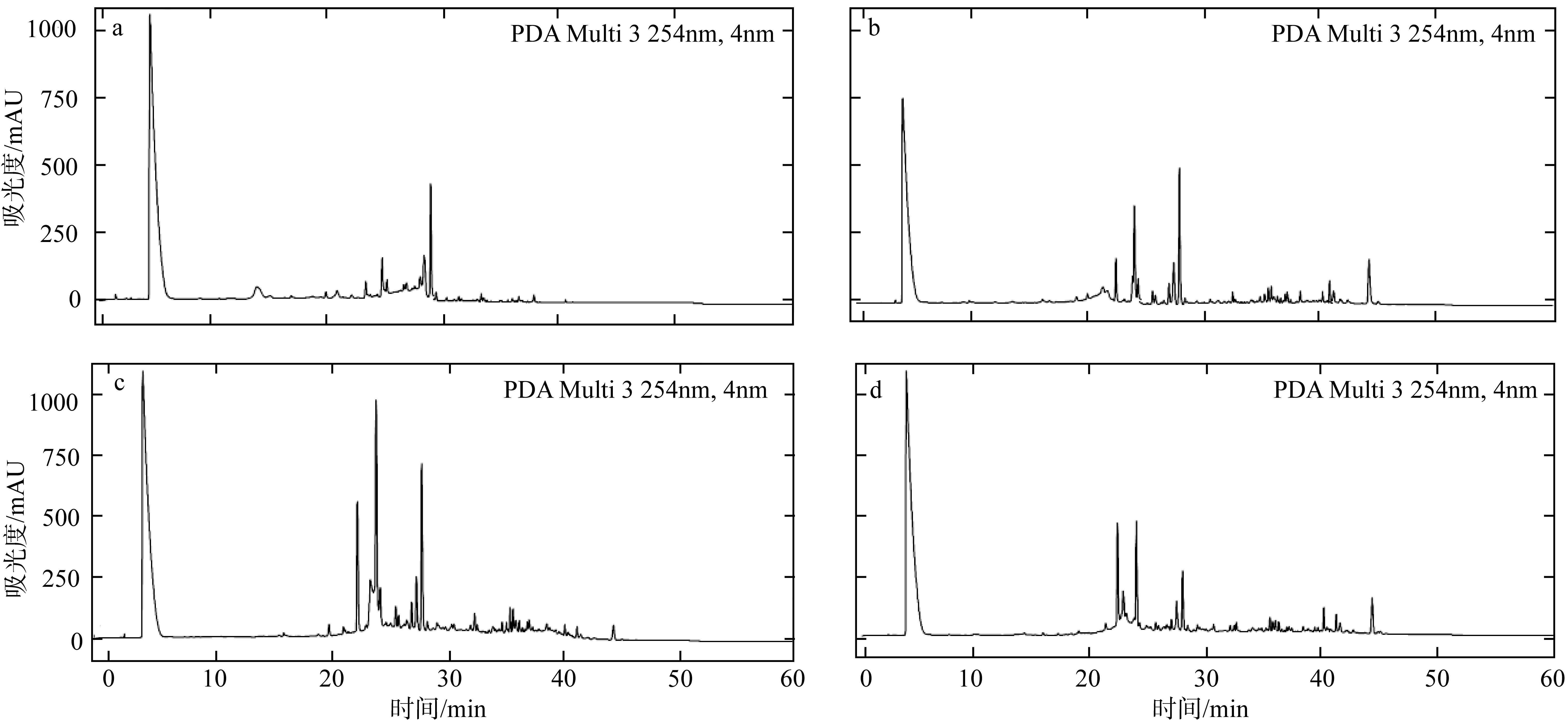
Molecular identification, secondary metabolites and biological activities of a deep-sea-derived fungus 101#*
ZENG Buyan1( ), LIANG Zhifeng1, LUO Qinqin3, ZENG Ling3, YANG Changgeng1, WANG Liyun1, SUN Yulin1,2(
), LIANG Zhifeng1, LUO Qinqin3, ZENG Ling3, YANG Changgeng1, WANG Liyun1, SUN Yulin1,2( )
)
- 1. School of Life Science and Technology, Lingnan Normal University, Zhanjiang 524048, China
2. Mangrove Institute, Lingnan Normal University, Zhanjiang 524048, China
3. School of Chemistry and Chemical Engineering, Lingnan Normal University, Zhanjiang 524048, China
-
Received:2022-11-11Revised:2022-12-26Online:2023-09-10Published:2022-12-28 -
Supported by:National Natural Science Foundation of China(31902373); Natural Science Foundation of Guangdong Province, China(2021A1515011398); Project of Education Department of Guangdong Province, China(2021ZDJS034); Project of Education Department of Guangdong Province, China(2021KCXTD039); Project of Education Department of Guangdong Province, China(2022-K08); Public Service Platform of Biomedical Resources Research and Development of South China Sea(2017C8B2); Yanling Outstanding Young Teacher Training Program of Lingnan Normal University(YL20200213); Open Project of Mangrove Research Institute, Lingnan Normal University(PYXM05)
Cite this article
ZENG Buyan, LIANG Zhifeng, LUO Qinqin, ZENG Ling, YANG Changgeng, WANG Liyun, SUN Yulin. Molecular identification, secondary metabolites and biological activities of a deep-sea-derived fungus 101#*[J].Journal of Tropical Oceanography, 2023, 42(5): 104-114.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Minimum inhibitory concentration (MIC) of crude extract of strain 101# against 12 indicator bacteria (n=3)"
| 指示菌 | 阳性对照 | 粗提物浓度/(μg·mL-1) | MIC/(μg·mL-1) | |||||
|---|---|---|---|---|---|---|---|---|
| 12.5 | 6.25 | 3.125 | 1.563 | 0.781 | 0.391 | |||
| 枯草芽孢杆菌 (Bacillus subtilis) | ++++ | ++ | ++ | + | + | + | - | 0.781 |
| 藤黄微球菌 (Micrococcus luteus) | ++++ | ++ | ++ | + | + | - | - | 1.563 |
| 苏云金芽孢杆菌 (Bacillus thuringiensis) | ++++ | ++ | + | + | + | - | - | 1.563 |
| 耐甲氧西林金黄色葡萄球菌 (Methicillin-resistant staphylococcus aureus) | ++++ | ++ | + | + | - | - | - | 3.125 |
| 金黄色葡萄球菌 (Staphylococcus aureus) | ++++ | ++ | + | + | - | - | - | 3.125 |
| 无乳链球菌 (Streptococcus agalactiae) | ++++ | ++ | + | - | - | - | - | 6.250 |
| 稀瓦氏菌 (Shewanella) | ++++ | ++ | + | + | + | - | - | 1.563 |
| 大肠杆菌 (Escherichia coli) | ++++ | ++ | + | + | - | - | - | 3.125 |
| 福氏志贺菌 (Shigella Flexneri) | ++++ | ++ | + | + | - | - | - | 3.125 |
| 肺炎克雷伯菌 (Klebsiella pneumoniae) | ++++ | + | + | - | - | - | - | 6.250 |
| 麦氏交替单胞菌 (Alteromonas macleodii) SCAU-003 | ++++ | +++ | ++ | + | - | - | - | 3.125 |
| 多耐白色念珠菌 (Candida albicans) | ++++ | +++ | ++ | + | + | - | - | 1.563 |
Tab. 2
Minimum inhibitory concentration (MIC) of crude extract of strain 101# against 6 plant pathogens (n=3)"
| 指示菌 | 阳性对照 | 粗提物浓度/(μg·mL-1) | MIC/(μg·mL-1) | ||||
|---|---|---|---|---|---|---|---|
| 200 | 100 | 50 | 25 | 12.5 | |||
| 禾谷镰刀菌 (Fusarium graminearum) | ++ | ++ | + | + | - | - | 50 |
| 意大利青霉 (Penicillium italicum) | ++++ | + | - | - | - | - | 200 |
| 尖孢镰刀菌 (Fusarium oxysporum) | + | + | - | - | - | - | 200 |
| 层出镰刀菌 (Fusarium proliferatum) | + | - | - | - | - | - | >200 |
| 盘长孢状刺盘孢 (Colletotrichum gloeosprioides) | + | - | - | - | - | - | >200 |
| 茄镰孢菌 (Fusarium solani) | ++ | - | - | - | - | - | >200 |
Tab 3.
Test results of lethal activity of strain 101# crude extract against brine shrimp"
| 时间/h | 粗提物浓度/( mg·mL-1) | LD50/(mg·mL-1) | 95%置信区间 | |||||
|---|---|---|---|---|---|---|---|---|
| 100 | 50 | 25 | 12.5 | 6.25 | 下限 | 上限 | ||
| 1 | 64.7 | 38.9 | 25.0 | 10.0 | 0.0 | 74.597 | 54.900 | 118.483 |
| 12 | 100.0 | 83.3 | 56.3 | 30.0 | 12.5 | 24.322 | 17.238 | 34.071 |
| 24 | 100.0 | 100.0 | 87.5 | 35.5 | 18.8 | 13.797 | 9.739 | 19.393 |
| 48 | 100.0 | 100.0 | 93.8 | 65.0 | 37.5 | 8.559 | 4.888 | 12.455 |
| [1] |
安昶亮, 2019. 海洋真菌Aspergillus sp. SCS-KFD66的次级代谢产物及其活性研究[D]. 南京: 南京农业大学:1-121.
|
|
|
|
| [2] |
董怡飞, 徐秀丽, 张新婉, 等, 2022. 深海真菌Aspergillus sp.20220129的次级代谢产物及抑菌活性研究[J]. 中国抗生素杂志, 47(5): 494-500.
|
|
|
|
| [3] |
黄智, 王发左, 田新朋, 等, 2012. 一株南海北部深海沉积环境真菌00457的鉴定及其活性研究[J]. 生物技术通报, (10): 199-204.
|
|
|
|
| [4] |
江北, 吕梦霞, 蒋冬花, 2019. 曲霉属真菌活性代谢产物及在农业生产中的应用研究进展[J]. 微生物学杂志, 39(2): 103-110.
|
|
|
|
| [5] |
罗寒, 李晓栋, 李晓明, 等, 2017. 红树林来源内生真菌杂色曲霉Aspergillus versicolor MA-229次级代谢产物研究[J]. 中国抗生素杂志, 42(4): 334-340.
|
|
|
|
| [6] |
潘力, 崔翠, 王斌, 2010. 一种用于PCR扩增的丝状真菌DNA快速提取方法[J]. 微生物学通报, 37(3): 450-453.
|
|
|
|
| [7] |
钱生辉, 2022. 南海红树林底泥海洋放线菌抗菌/抗群体感应活性菌株的筛选及次级代谢产物的初步研究[D]. 扬州: 扬州大学: 1-90.
|
|
|
|
| [8] |
单体壮, 2020. 海洋真菌Aspergillus ruber的次级代谢产物及其生物活性研究[D]. 扬州: 扬州大学: 1-137.
|
|
|
|
| [9] |
谭雁鸿, 李基兴, 林秀萍, 等, 2019. 中国南海软珊瑚真菌Eupenicillium sp. DX-SER3 (KC871024) 的次级代谢产物研究[J]. 热带海洋学报, 38(2): 43-47.
doi: 10.11978/2018072 |
|
|
|
| [10] |
王发左, 2008. 海洋真菌抗肿瘤活性次级代谢产物及其生物转化研究[D]. 青岛: 中国海洋大学: 21-22.
|
|
|
|
| [11] |
叶禹秀, 罗小卫, 杨斌, 等, 2022. 南海礁栖海藻共附生真菌Pestalotiopsis neglecta SCSIO41 403次级代谢产物研究[J]. 热带海洋学报, 41(3): 186-190.
doi: 10.11978/2021089 |
|
|
|
| [12] |
于清武, 胡丽琴, 李菲, 等, 2015. 南海深海沉积物可培养细菌多样性及其生物毒性分析[J]. 南方农业学报, 46(12): 2203-2208.
|
|
|
|
| [13] |
曾奇, 仲伟茂, 向瑶, 等, 2018. 南海深海沉积物中52株真菌的初步分离鉴定及其代谢产物活性[J]. 微生物学通报, 45(9): 1904-1915.
|
|
|
|
| [14] |
张长生, 李文利, 2018. 海洋微生物学: 新机遇, 新挑战[J]. 微生物学通报, 49(5): 1841-1842.
|
|
|
|
| [15] |
|
| [16] |
doi: 10.1039/c7np00052a pmid: 29335692 |
| [17] |
doi: 10.1039/d0np00089b pmid: 33570537 |
| [18] |
doi: 10.1039/D1NP00076D |
| [19] |
doi: 10.1021/acs.jnatprod.1c00776 |
| [20] |
pmid: 15921427 |
| [21] |
doi: 10.1080/14786419.2018.1480622 |
| [22] |
doi: 10.3390/md17050262 |
| [23] |
doi: 10.1016/S0040-4039(00)84436-6 |
| [24] |
doi: 10.1021/jm00275a029 |
| [25] |
doi: 10.1021/acs.jnatprod.9b01285 pmid: 32162523 |
| [26] |
doi: 10.3390/antibiotics10081010 |
| [27] |
doi: 10.3390/md18030164 |
| [28] |
|
| [29] |
doi: 10.1016/j.phytochem.2021.112967 |
| [30] |
doi: 10.1007/s10600-020-02955-x |
| [31] |
doi: 10.1080/14786419.2018.1465423 |
| [1] | AN Fan, JIANG Yue, WANG Yu, CAO Guangping, GAO Chenghai, LIU Yonghong, YI Xiangxi, BAI Meng. Studies on secondary metabolites of endophytic fungus Aspergillus terreus GXIMD 03158 isolated from mangroves Acanthus ilicifolius L. [J]. Journal of Tropical Oceanography, 2024, 43(5): 41-48. |
| [2] | SUN Manman, ZENG Yanbo, XU Han, YAO Ligong, GUO Yuewei, SU Mingzhi. Chemical composition and antibacterial activities of the soft coral Lobophytum sp. from the South China Sea [J]. Journal of Tropical Oceanography, 2024, 43(4): 189-197. |
| [3] | LIN Yingting, SUN Ying, HU Jiangnan, DONG Shuai. Study on secondary metabolites of the endophytic fungus Talaromyces sp. XXH006 Isolated from Conus literatus [J]. Journal of Tropical Oceanography, 2024, 43(4): 181-188. |
| [4] | WANG Jiaxi, LU Humu, QI Xin, GAO Chenghai, LIU Yonghong, LUO Xiaowei. Study on the secondary metabolites from the Weizhou Island coral Acropora austera associated fungus Arachniotus ruber GXIMD 02510 and their antibacterial activity [J]. Journal of Tropical Oceanography, 2024, 43(4): 174-180. |
| [5] | QUAN Miaomiao, MA Qingyun, YANG Li, XIE Qingyi, DAI Haofu, HAO Yu’e, ZHAO Youxing. Study on the secondary metabolites of marine fungus Phaeospheriopsis sp. ZYX-Z-811 [J]. Journal of Tropical Oceanography, 2024, 43(2): 121-127. |
| [6] | LIU Ying, XIAO Yang, ZHU Zhenxin, LIU Hongcun, JIANG Mingguo, ZHU Yuzhang, LIN Kun, WU Jincheng, LU Xiaomei, HUANG Xiaoning, LIANG Haina, LU Wensen, YANG Lifang. Study on the Secondary Metabolites of Marine Streptomyces Sporoverrucosus 33510 [J]. Journal of Tropical Oceanography, 2024, 43(2): 128-134. |
| [7] | FENG Guangfu, HE Jeyi, BAI Meng, YI Xiangxi, LIU Xinming, LUO Lianxiang, GAO Chenghai. Secondary Metabolites from Hydaranth of Coral Goniopora columna in the Beibu Gulf [J]. Journal of Tropical Oceanography, 2024, 43(2): 135-139. |
| [8] | XING Nannan, REN Runxin, TANG Zhenzhou, LUO Zhihong, XIA Chenxi, LIU Yonghong, PENG Liang, CHEN Xianqiang. Study on the secondary metabolites of fungus Aspergillus sp. GXIMD02003 derived from marine sediment in the Weizhou island [J]. Journal of Tropical Oceanography, 2023, 42(5): 154-160. |
| [9] | LIANG Hanqiao, CHEN Wenfeng, FAN Yikai, ZHU Zidong, MA Guoxu, CHEN Deli, TIAN Jing. Research progress on the secondary metabolites and activities of endophytic fungi of genus Aspergillus and Trichoderma from mangroves [J]. Journal of Tropical Oceanography, 2023, 42(4): 12-24. |
| [10] | LU Tianmei, GUAN jiasong, QIN Shijing, LIU Yonghong, SU Zhiwei. Preliminary study on the culturable myxobacteria resources from the Beibu Gulf, Guangxi and their antibacterial activity [J]. Journal of Tropical Oceanography, 2023, 42(3): 158-168. |
| [11] | YE Yuxiu, LUO Xiaowei, YANG Bin, LIN Xiuping, LIU Yonghong, ZHOU Xuefeng. Study on the secondary metabolites of reef habitat algae-derived fungus Pestalotiopsis neglecta SCSIO41403 from the South China Sea [J]. Journal of Tropical Oceanography, 2022, 41(3): 186-190. |
| [12] | ZHANG Han, TAN Yanhong, YANG Bin, LIU Yonghong, LI Yunqiu. Study on the secondary metabolites from the South China Sea soft coral-derived fungus Acremonium sp. SCSIO41216 [J]. Journal of Tropical Oceanography, 2021, 40(6): 135-139. |
| [13] | Xiji XIAO,Yun DENG,Lufeng XIE,Qi ZHANG,Xia WEI,Xin YU,Cuixian ZHANG. Research on Sterols from Leptogorgia rigida [J]. Journal of Tropical Oceanography, 2019, 38(4): 64-69. |
| [14] | Yanhong TAN, Jixing LI, Xiuping LIN, Bin YANG, Yonghong LIU, Yunqiu LI. Study on the secondary metabolites from the South China Sea soft coral-derived fungus Eupenicillium sp. DX-SER3 (KC871024) [J]. Journal of Tropical Oceanography, 2019, 38(2): 43-47. |
| [15] | JIANG Xiao,REN Chun-hua,HU Chao-qun,LUO Peng,CHEN Chang,FENG Jing-bi. On dynamic change of Vibrio species in the Daya Bay using molecular identification method [J]. Journal of Tropical Oceanography, 2010, 29(4): 154-159. |
|
||