Journal of Tropical Oceanography ›› 2021, Vol. 40 ›› Issue (4): 22-34.doi: 10.11978/2020067CSTR: 32234.14.2020067
• Marine Biology • Previous Articles Next Articles
Physiological and genomic characteristics of two lytic phages infecting Vibrio alginolyticus
ZHONG Wanxuan1,2( ), YANG Yunlan3, LI Xiangfu1,4, XU Jie1,2,4(
), YANG Yunlan3, LI Xiangfu1,4, XU Jie1,2,4( )
)
- 1. State Key Laboratory of Tropical Oceanography, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
2. College of Marine Science, University of Chinese Academy of Sciences, Beijing 100049, China
3. College of the Environment and Ecology, State Key Laboratory for Marine Environmental Science, Xiamen University, Xiamen, 361102 China
4. Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou, 510301 China
-
Received:2020-06-24Revised:2020-07-26Online:2021-07-10Published:2020-07-29 -
Contact:XU Jie E-mail:zhongwanxuan17@mails.ucas.ac.cn;xujie@scsio.ac.cn -
Supported by:Project of Program of Science and Technology of Guangzhou(20190420029);National Natural Science Foundation of China(41506095);National Natural Science Foundation of China(41676075);Project of Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences(ISEE2019ZR02);Project of Department of Science and Technology of Guangdong Province(2018B030320005);China Postdoctoral Science Foundation(2019M662237)
CLC Number:
- P735.531
Cite this article
ZHONG Wanxuan, YANG Yunlan, LI Xiangfu, XU Jie. Physiological and genomic characteristics of two lytic phages infecting Vibrio alginolyticus[J].Journal of Tropical Oceanography, 2021, 40(4): 22-34.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks

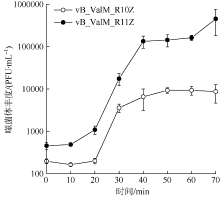
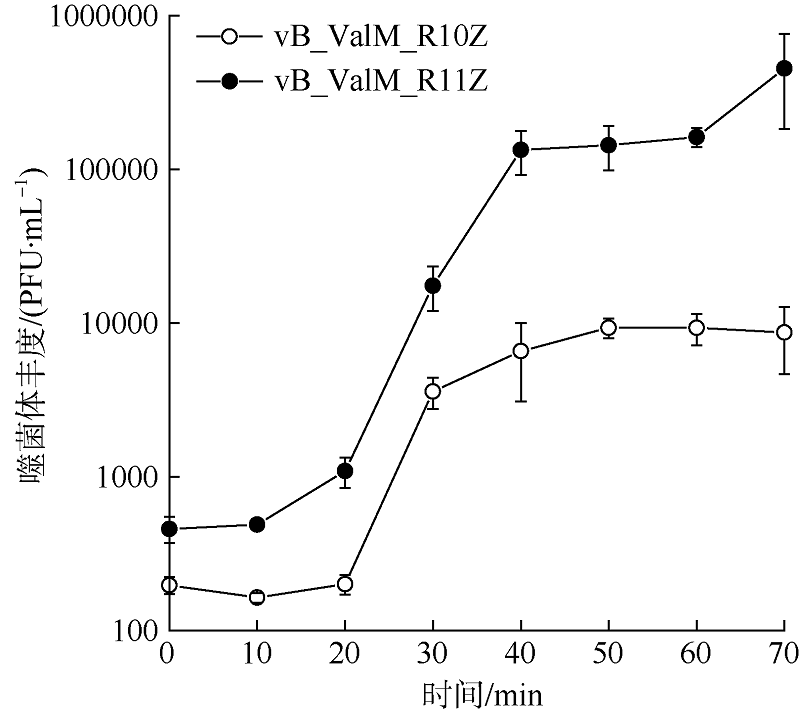
Fig. 1
Phage plaques on double-layer plate and morphology in transmission electron microscopy of phages vB_ValM_R10Z and vB_ValM_R11Z a. Phage plaques of vB_ValM_R10Z; b. phage plaques of vB_ValM_R11Z; c. transmission electron microscopy images of vB_ValM_R10Z; d. transmission electron microscopy images of vB_ValM_R11Z"

Tab. 1
The host ranges of vB_ValM_R10Z and vB_ValM_R11Z"
| 细菌种属 | 株系 | 致病性 | vB_ValM_R10Z | vB_ValM_R11Z |
|---|---|---|---|---|
| V. alginolyticus | ATCC 17749T | 可致病 | + | + |
| V. alginolyticus | JL2674 | 未知 | - | - |
| V. campbellii | JL2671 | 未知 | - | - |
| V. campbellii | JL3506 | 未知 | - | - |
| V. parahemolyticus | ATCC 17802T | 可致病 | - | - |
| V. parahemolyticus | BVP2 | 未知 | - | - |
| V. parahemolyticus | 20160623-13 | 可致病 | - | - |
| V. parahemolyticus | 20160719-2 | 可致病 | - | - |
| V. parahemolyticus | 20160615-5 | 可致病 | - | - |
| V. harveyi | BVH1 | 可致病 | - | - |
| V. harveyi | 20150916-2 | 可致病 | - | - |
| V. owensii | JL2581 | 未知 | - | - |
| V. owensii | JL3186 | 未知 | + | + |
| V. owensii | JL3187 | 未知 | - | - |
| V. caribbeanicus | JL3219 | 未知 | - | - |
| V. fortis | JL3515 | 未知 | - | - |
| V. chagasii | JL3518 | 未知 | - | - |
| V. inhibens | JL3707 | 未知 | - | - |
| V. cholerae | 20161020-5 | 可致病 | - | - |
| V. variabilis | JL3833 | 未知 | - | - |
Tab. 2
tRNA genes of phages vB_ValM_R10Z and vB_ValM_R11Z"
| vB_ValM_R10Z | vB_ValM_R11Z | |||||||
|---|---|---|---|---|---|---|---|---|
| tRNA基因 序号 | 位置 | 氨基酸 | 反密码子 | tRNA基因 序号 | 位置 | 氨基酸 | 反密码子 | |
| 1 | 164151~164264 | Phe | AAA | 1 | 163682~163794 | Phe | AAA | |
| 2 | 164275~164399 | Asn | GTT | 2 | 163805~163929 | Asn | GTT | |
| 3 | 164784~164861 | Met | CAT | 3 | 164314~164391 | Met | CAT | |
| 4 | 166706~166779 | Pro | TGG | 4 | 166236~166309 | Pro | TGG | |
| 5 | 166789~166865 | Pro | TGG | 5 | 166319~166395 | Pro | TGG | |
| 6 | 166873~166948 | Arg | ACG | 6 | 166404~166479 | Arg | ACG | |
| 7 | 167302~167378 | Leu | TAG | 7 | 166833~166909 | Leu | TAG | |
| 8 | 167380~167454 | Phe | GAA | 8 | 166911~166985 | Phe | GAA | |
| 9 | 167544~167618 | Lys | TTT | 9 | 167075~167149 | Lys | TTT | |
| 10 | 167630~167706 | Lys | TTT | 10 | 167161~167237 | Lys | TTT | |
| 11 | 167770~167844 | Glu | TTC | 11 | 167301~167375 | Glu | TTC | |
| 12 | 167851~167936 | Tyr | GTA | 12 | 167382~167467 | Tyr | GTA | |
| 13 | 168039~168115 | Met | CAT | 13 | 167570~167646 | Met | CAT | |
| 14 | 168411~168509 | Ser | TGA | 14 | 167942~168040 | Ser | TGA | |
| 15 | 168574~168647 | Ile | GAT | 15 | 168105~168178 | Ile | GAT | |
| 16 | 168654~168727 | Trp | CCA | 16 | 168185~168258 | Trp | CCA | |
| 17 | 168803~168877 | Asn | GTT | 17 | 168334~168408 | Asn | GTT | |
| 18 | 168879~168954 | Asn | GTT | 18 | 168410~168485 | Asn | GTT | |
| 19 | 169024~169099 | Asp | GTC | 19 | 168554~168629 | Asp | GTC | |
| 20 | 169119~169192 | Gln | TTG | 20 | 168649~168722 | Gln | TTG | |
| 21 | 169322~169396 | Thr | TGT | 21 | 168850~168925 | Thr | TGT | |
| 22 | 169968~170042 | Ala | TGC | 22 | 169167~169241 | Ala | TGC | |
| 23 | 170117~170192 | Arg | TCT | 23 | 169316~169391 | Arg | TCT | |
| vB_ValM_R10Z | vB_ValM_R11Z | |||||||
| tRNA基因 序号 | 位置 | 氨基酸 | 反密码子 | tRNA基因 序号 | 位置 | 氨基酸 | 反密码子 | |
| 24 | 170717~170792 | Met | CAT | 24 | 169915~169990 | Met | CAT | |
| 25 | 170982~171055 | Gly | TCC | 25 | 170180~170253 | Gly | TCC | |
| 26 | 171062~171135 | Val | TAC | 26 | 170260~170333 | Val | TAC | |
| 27 | 171551~171627 | His | GTG | 27 | 170749~170825 | His | GTG | |
| 28 | 171640~171713 | Cys | GCA | 28 | 170838~-170911 | Cys | GCA | |
| 29 | 171716~171792 | Leu | TAA | 29 | 170914~170990 | Leu | TAA | |
| 30 | 171909~171985 | Leu | CAA | 30 | 171107~171183 | Leu | CAA | |
| 31 | 172154~172242 | Ser | GCT | 31 | 171352~171440 | Ser | GCT | |

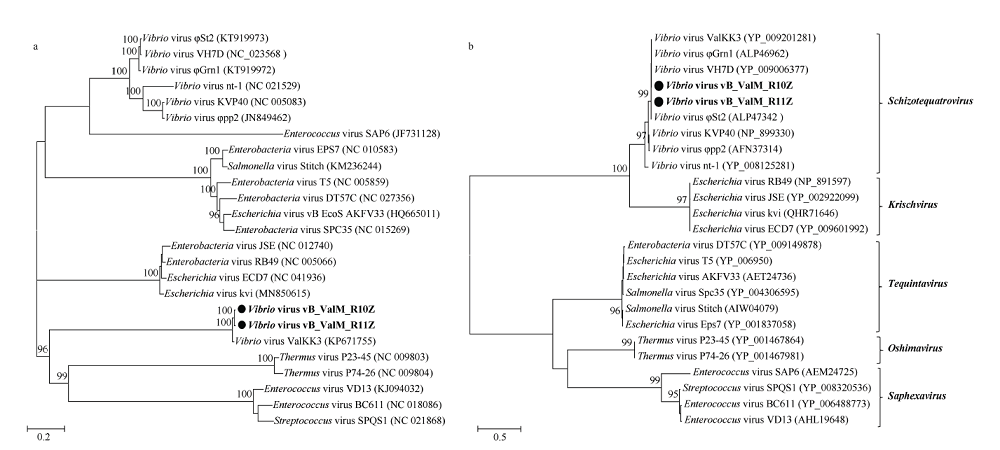
Fig. 4
Phy logenetic trees of complete genome (a) and DNA polymerase (b) of phages vB_ ValM_ R10Z, vB_ ValM_R11Z and other known phages Maximum likelihood analyses with 1000 bootstrap replicates were used to derive the trees based on amino-acid sequences. Bootstrap values are shown above the major nodes, and the values below 90 are hidden. Phages vB_ValM_R10Z and vB_ValM_R11Z are marked in bold. Numbers in brackets represent corresponding GenBank ID. The phylogenetic trees include phages vB_ValM_ R10Z and vB_ValM_ R11Z, as well as all published Schizotequatrovirus phages and some representative phages of other genera"
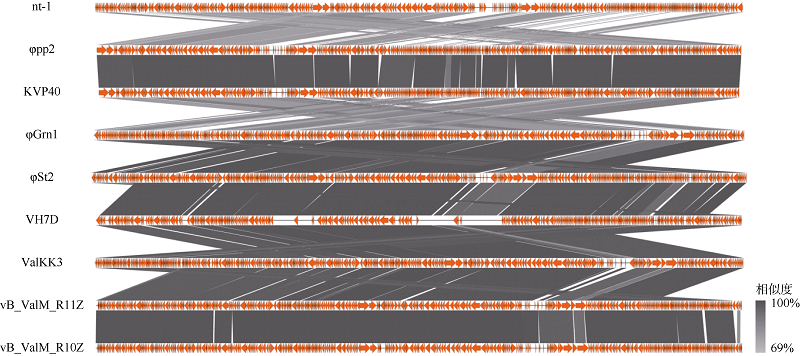
Tab. 3
Comparison of Schizotequatrovirus phages"
| 噬菌体 | vB_ValM_R10Z | vB_ValM_R11Z | VH7D | φSt2 | φGrn1 | ValKK3 | KVP40 | φpp2 | nt-1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 宿主菌 | V. alginolyticus | V. alginolyticus | V. sp. 7D | V. alginolyticus | V. alginolyticus | V. alginolyticus | V. parahaemolyticus | V. parahaemolyticus | V. natrigens | ||||||||||
| 地点 | 中国东山岛 | 中国东山岛 | 中国厦门 | 希腊克里特岛 | 希腊克里特岛 | 马来西亚 | 日本Urado湾 | 中国台湾南部 | 大西洋 | ||||||||||
| 分离环境 | 养殖水体 | 污水 | 养殖水体 | 海水 | 海水 | 沉积物 | 污水 | 养殖水体 | 沉积物 | ||||||||||
| 头长/nm | 147 | 148 | 155 | 151 | 138 | 140 | 140 | 150 | 120 | ||||||||||
| 头宽/nm | 78 | 81 | 75 | 81 | 74 | 79 | 70 | 90 | 70 | ||||||||||
| 尾长/nm | 120 | 114 | 132 | 134 | 120 | 130 | 110 | 110 | |||||||||||
| 潜伏期/min | 20 | 20 | 30 | 30 | |||||||||||||||
| 裂解量/PFU·cell-1 | 43 | 114 | 97 | 44 | |||||||||||||||
| 噬菌体 | vB_ValM_R10Z | vB_ValM_R11Z | VH7D | φSt2 | φGrn1 | ValKK3 | KVP40 | φpp2 | nt-1 | ||||||||||
| GenBank 收录号 | MT612988 | MT612989 | KC131129.1 | KT919973.1 | KT919972 | KP671755.1 | AY283928 | JN849462 | HQ317393 | ||||||||||
| 基因组大小/bp | 247167 | 246831 | 246964 | 250485 | 248605 | 248088 | 244835 | 246421 | 247489 | ||||||||||
| G+C值 | 41.30% | 41.33% | 41.31% | 42.60% | 38.80% | 41.20% | 42.60% | 42.55% | 41.33% | ||||||||||
| ORF数量/个 | 386 | 385 | 387 | 412 | 410 | 390 | 386 | 383 | 379 | ||||||||||
| tRNA数量/个 | 31 | 31 | 27 | 29 | 28 | 30 | 30 | 28 | |||||||||||
| 毒力基因 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | ||||||||||
| 抗药性基因 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | 无 | ||||||||||
| 参考文献 | 本文 | 本文 |
| [1] | 冯书章, 刘军, 孙洋, 2007. 细菌的病毒——噬菌体最新分类与命名[J]. 中国兽医学报, 27(4):604-608 (in Chinese). |
| [2] | 黄瑞芳, 2005. 斜带石斑鱼溶藻弧菌病的研究[J]. 水产科学, 24(6):1-3. |
| HUANG RUIFANG, 2005. Vibrio alginolyticus in the Grouper, Epinephelus coioides[J]. Fisheries Science, 24(6):1-3 (in Chinese with English abstract). | |
| [3] | 李自强, 2018. 哈维氏弧菌噬菌体的分离及其生物学特性[D]. 厦门: 厦门大学: 41-42. |
| LI ZIQIANG, 2018. Isolation and biological characteristics of Vibrio harveyi bacteriophage[D]. Xiamen: Xiamen University: 41-42 (in Chinese with English abstract). | |
| [4] | 刘淇, 王学忠, 戴芳钰, 等, 2007. 梭子蟹溶藻弧菌病的初步研究[J]. 齐鲁渔业, 24(9):1-3. |
| LIU QI, WANG XUEZHONG, DAI FANGYU, et al, 2007. Preliminary study on Vibrio alginolyticus disease in Portunus trituberculatus[J]. Shandong Fisheries, 24(9):1-3 (in Chinese with English abstract). | |
| [5] | 农业农村部渔业渔政管理局, 全国水产技术推广总站, 中国水产学会, 2019. 中国渔业统计年鉴-2019[M]. 北京: 中国农业出版社: 130-132. |
| MARA Bureau of Fisheries, National Fisheries Technology Extension Center, China Society of Fisheries, 2019. 2019 China fishery statistical yearbook[M]. Beijing: China Agriculture Press: 130-132(in Chinese). | |
| [6] | 张晓华, 林禾雨, 孙浩, 2018. 弧菌科分类学研究进展[J]. 中国海洋大学学报, 48(8):43-56. |
| ZHANG XIAOHUA, LIN HEYU, SUN HAO, 2018. Taxonomy of vibrionaceae: a review[J]. Periodical of Ocean University of China, 48(8):43-56 (in Chinese with English abstract). | |
| [7] | 张昕, 蔡俊鹏, 2004. 噬菌体在水产养殖中的应用[J]. 粮食与饲料工业, (6):38-39. |
| ZHANG XIN, CAI JUNPENG, 2004. Application of phages in aquaculture[J]. Cereal & Feed Industry, (6):38-39 (in Chinese). | |
| [8] | 张正福, 纪来升, 1981. 密度梯度离心技术在分子生物学研究中的应用[J]. 植物生理学通讯, (6):60-64. |
| ZHANG ZHEN GFU, JI LAISHENG, 1981. Application of density gradient centrifugation technique in molecular biology research[J]. Plant Physiology Journal, (6):60-64 (in Chinese). | |
| [9] | ALCOCK B P, RAPHENYA A R, LAU T T Y, et al, 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database[J]. Nucleic Acids Res, 48(D1):D517-D525. |
| [10] |
AL-SHAYEB B, SACHDEVA R, CHEN LINXING, et al, 2020. Clades of huge phages from across Earth's ecosystems[J]. Nature, 578(7795):425-431.
doi: 10.1038/s41586-020-2007-4 |
| [11] |
BOLGER A M, LOHSE M, USADEL B, 2014. Trimmomatic: a flexible trimmer for Illumina sequence data[J]. Bioinformatics, 30(15):2114-2120.
doi: 10.1093/bioinformatics/btu170 |
| [12] |
CHEN LIHONG, YANG JIAN, YU JUN, et al, 2005. VFDB: a reference database for bacterial virulence factors[J]. Nucleic Acids Res, 33(S1):D325-D328.
doi: 10.1093/nar/gki008 |
| [13] |
CHEN LING, FAN JIQIANG, YAN TINGWEI, et al, 2019. Isolation and Characterization of Specific Phages to Prepare a Cocktail Preventing Vibrio sp. Va-F3 Infections in Shrimp (Litopenaeus vannamei)[J]. Frontiers in Microbiology, 10:2337.
doi: 10.3389/fmicb.2019.02337 |
| [14] |
CRUMMETT L T, PUXTY R J, WEIHE C, et al, 2016. The genomic content and context of auxiliary metabolic genes in marine cyanomyoviruses[J]. Virology, 499:219-229.
doi: 10.1016/j.virol.2016.09.016 |
| [15] |
EGIDIUS E, 1987. Vibriosis: pathogenicity and pathology. A review[J]. Aquaculture, 67(1-2):15-28.
doi: 10.1016/0044-8486(87)90004-4 |
| [16] | GUERRERO-FERREIRA R C, VIOLLIER P H, ELY B, et al, 2011. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus[J]. Proceedings of the National Academy of Sciences of the United States of America, 108(24):9963-9968. |
| [17] |
HUTINET G, KOT W, CUI L, et al, 2019. 7-Deazaguanine modifications protect phage DNA from host restriction systems[J]. Nature Communications, 10:5442.
doi: 10.1038/s41467-019-13384-y |
| [18] |
INA-SALWANY M Y, AL-SAARI N, MOHAMAD A, et al, 2019. Vibriosis in fish: a review on disease development and prevention[J]. Journal of Aquatic Animal Health, 31(1):3-22.
doi: 10.1002/aah.2019.31.issue-1 |
| [19] |
KALATZIS P G, BASTIAS R, KOKKARI C, et al, 2016. Isolation and characterization of two lytic bacteriophages, φSt2 and φGrn1; phage therapy application for biological control of Vibrio alginolyticus in aquaculture live feeds[J]. PLoS One, 11(3):e0151101.
doi: 10.1371/journal.pone.0151101 |
| [20] |
KALATZIS P G, CASTILLO D, KATHARIOS P, et al, 2018. Bacteriophage interactions with marine pathogenic vibrios: implications for phage therapy[J]. Antibiotics, 7(1):15.
doi: 10.3390/antibiotics7010015 |
| [21] |
KILJUNEN S, HAKALA K, PINTA E, et al, 2005. Yersiniophage ϕR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine[J]. Microbiology, 151(12):4093-4102.
doi: 10.1099/mic.0.28265-0 |
| [22] |
LAL T M, SANO M, HATAI K, et al, 2016. Complete genome sequence of a giant Vibrio phage ValKK3 infecting Vibrio alginolyticus[J]. Genomics Data, 8:37-38.
doi: 10.1016/j.gdata.2016.03.002 |
| [23] |
LIN YINGRONG, LIN C-S, 2012. Genome-wide characterization of Vibrio phage ϕpp2 with unique arrangements of the mob-like genes[J]. BMC Genomics, 13:244.
doi: 10.1186/1471-2164-13-244 |
| [24] |
LIU WANGTA, LIN YINGRONG, LU MINGWEI, et al, 2014. Genome sequences characterizing five mutations in RNA polymerase and major capsid of phages ϕA318 and ϕAs51 of Vibrio alginolyticus with different burst efficiencies[J]. BMC Genomics, 15:505.
doi: 10.1186/1471-2164-15-505 |
| [25] | LIU XIAOFEI, CAO YUAN, ZHANG HELIN, et al, 2015. Complete genome sequence of Vibrio alginolyticus ATCC 17749T[J]. Genome Announc, 3(1):e01500-14. |
| [26] |
LOWE T M, CHAN P P, 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes[J]. Nucleic Acids Research, 44(W1):W54-W57.
doi: 10.1093/nar/gkw413 |
| [27] |
LU LONGFEI, CAI LANLAN, JIAO NIANZHI, et al, 2017. Isolation and characterization of the first phage infecting ecologically important marine bacteria Erythrobacter[J]. Virology Journal, 14:104.
doi: 10.1186/s12985-017-0773-x |
| [28] |
LUO ZHUHUA, YU YANPING, JOST G, et al, 2015. Complete genome sequence of a giant Vibrio bacteriophage VH7D[J]. Marine Genomics, 24:293-295.
doi: 10.1016/j.margen.2015.10.005 |
| [29] |
LV TENGTENG, SONG TONGXIANG, LIU HUIJIE, et al, 2019. Isolation and characterization of a virulence related Vibrio alginolyticus strain Wz11 pathogenic to cuttlefish, Sepia pharaonis[J]. Microbial Pathogenesis, 126:165-171.
doi: 10.1016/j.micpath.2018.10.041 |
| [30] |
MAK A N S, LAMBERT A R, STODDARD B L, 2010. Folding, DNA recognition, and function of GIY-YIG endonucleases: crystal structures of R.Eco29kI[J]. Structure, 18(10):1321-1331.
doi: 10.1016/j.str.2010.07.006 |
| [31] |
MATSUZAKI S, TANAKA S, KOGA T, et al, 1992. A broad-host-range Vibriophage, KVP40, isolated from sea water[J]. Microbiology and Immunology, 36(1):93-97.
doi: 10.1111/mim.1992.36.issue-1 |
| [32] | MIDDELBOE M, CHAN A M, BERTELSEN S K, 2010. Isolation and life-cycle characterization of lytic viruses infecting heterotrophic bacteria and cyanobacteria[M]// WILHELM S W, WEINBAUER M G, SUTTLE C A. Manual of aquatic viral ecology. Waco, Texas: American Society of Limnology and Oceanography: 118-133. |
| [33] |
MILLER E S, HEIDELBERG J F, EISEN J A, et al, 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage[J]. Journal of Bacteriology, 185(17):5220-5233.
doi: 10.1128/JB.185.17.5220-5233.2003 |
| [34] |
MOHAMAD N, AMAL M N A, YASIN I S M, et al, 2019. Vibriosis in cultured marine fishes: a review[J]. Aquaculture, 512:734289.
doi: 10.1016/j.aquaculture.2019.734289 |
| [35] |
NAKAI T, PARK S C, 2002. Bacteriophage therapy of infectious diseases in aquaculture[J]. Research in Microbiology, 153(1):13-18.
doi: 10.1016/S0923-2508(01)01280-3 |
| [36] | O'DONNELL M, LANGSTON L, STILLMAN B, 2013. Principles and concepts of DNA replication in bacteria, archaea, and eukarya[J]. Cold Spring Harbor Perspectives in Biology, 5(7):a010108. |
| [37] |
PÉREZ-SÁNCHEZ T, MORA-SÁNCHEZ B, BALCÁZAR J L, 2018. Biological approaches for disease control in aquaculture: advantages, limitations and challenges[J]. Trends in Microbiology, 26(11):896-903.
doi: 10.1016/j.tim.2018.05.002 |
| [38] |
SCHMIDT U, CHMEL H, COBBS C, 1979. Vibrio alginolyticus infections in humans[J]. Journal of Clinical Microbiology, 10(5):666-668.
doi: 10.1128/jcm.10.5.666-668.1979 |
| [39] |
SHARMA S, CHATTERJEE S, DATTA S, et al, 2017. Bacteriophages and its applications: an overview[J]. Folia Microbiologica, 62(1):17-55.
doi: 10.1007/s12223-016-0471-x |
| [40] | SKLIROS D, KALATZIS P G, KATHARIOS P, et al, 2016. Comparative functional genomic analysis of two Vibrio phages reveals complex metabolic interactions with the host cell[J]. Frontiers in Microbiology, 7:1807. |
| [41] |
STALIN N, SRINIVASAN P, 2017. Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India[J]. Veterinary Microbiology, 207:83-96.
doi: 10.1016/j.vetmic.2017.06.006 |
| [42] |
SULLIVAN M B, HUANG K H, IGNACIO-ESPINOZA J C, et al, 2010. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments[J]. Environmental Microbiology, 12(11):3035-3056.
doi: 10.1111/emi.2010.12.issue-11 |
| [43] |
WILSON J H 1973. Function of the bacteriophage T4 transfer RNA's[J]. Journal of Molecular Biology, 74(4): 753-754, IN27-IN28, 755-757.
doi: 10.1016/0022-2836(73)90065-X |
| [44] |
XIE JIASONG, BU LINGFEI, JIN SHAN, et al, 2020. Outbreak of vibriosis caused by Vibrio harveyi and Vibrio alginolyticus in farmed seahorse Hippocampus kuda in China[J]. Aquaculture, 523:735168.
doi: 10.1016/j.aquaculture.2020.735168 |
| [45] |
YANG YUNLAN, CAI LANLAN, MA RUIJIE, et al, 2017. A novel roseosiphophage isolated from the oligotrophic South China Sea[J]. Viruses, 9(5):109.
doi: 10.3390/v9050109 |
| [46] | YUAN YIHUI, GAO MEIYING, 2017. Jumbo bacteriophages: an overview[J]. Frontiers in Microbiology, 8:403. |
| [47] |
ZACHARY A, 1976. Physiology and ecology of bacteriophages of the marine bacterium Beneckea natriegens: salinity[J]. Applied and Environmental Microbiology, 31(3):415-422.
doi: 10.1128/aem.31.3.415-422.1976 |
| [48] |
ZENG QINGLU, BONOCORA R P, SHUB D A 2009. A free-standing homing endonuclease targets an intron insertion site in the psbA gene of cyanophages[J]. Current Biology, 19(3):218-222.
doi: 10.1016/j.cub.2008.11.069 |
| [49] |
ZERBINO D R, BIRNEY E, 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs[J]. Genome Research, 18(5):821-829.
doi: 10.1101/gr.074492.107 |
| [50] |
ZHANG JIANCHENG, CAO ZHENHUI, LI ZHEN, et al, 2015. Effect of bacteriophages on Vibrio alginolyticus infection in the sea cucumber, Apostichopus japonicus (Selenka)[J]. Journal of the World Aquaculture Society, 46(2):149-158.
doi: 10.1111/jwas.12177 |
| [1] | HUO Jiaxin, LI Yingxin, SONG Yan, ZHU Qing, ZHOU Weihua, YUAN Xiangcheng, HUANG Hui, LIU Sheng. Complete mitochondrial genome of Cladopsammia gracilis and Rhizopsammia wettsteini (Scleractinia, Dendrophylliidae) and its phylogenetic implications* [J]. Journal of Tropical Oceanography, 2024, 43(3): 22-30. |
| [2] | SUN Cuici, YUE Weizhong, ZHAO Wenjie, WANG Youshao. Distribution of the microbial Carbohydrate-Active enzymes genes in the surface sediment of the Daya Bay, China [J]. Journal of Tropical Oceanography, 2023, 42(5): 76-91. |
| [3] | HUANG Peixian, YAO Xuemei, YU Qiaochi, ZHANG Jiayu. Mitogenome characteristics and phylogenetic analysis of Siphonosoma australe in Hainan [J]. Journal of Tropical Oceanography, 2023, 42(2): 54-63. |
| [4] | LIU Wei, GUO Haipeng, DONG Pengsheng, YAN Mengchen, ZHANG Demin. Draft genome sequence and comparative genome analysis of Alliroseovarius sp. Z3 [J]. Journal of Tropical Oceanography, 2022, 41(1): 52-61. |
| [5] | ZHENG Chengzhi, ZUO Liming, MA Wang, ZHU Qin, WANG Huohuo, LÜ Songhui, CHEN Heng, HUANG Kaixuan. Interactions among Aureococcus anophagefferens, Skeletonema costatum, and Chattonella marina under different temperatures [J]. Journal of Tropical Oceanography, 2021, 40(3): 124-131. |
| [6] | CHEN Yao, YANG Xilu, HE Xuejia. Grazing of three common protozoan on brown tide alga Aureococcus anophagefferens [J]. Journal of Tropical Oceanography, 2018, 37(6): 120-132. |
| [7] | Youyun LU, Ming XUE, Zhihua LI, Chongqing WEN. Whole-genome sequencing and analysis of marine Bdellovibrio-and-like organism DA5 [J]. Journal of Tropical Oceanography, 2018, 37(6): 112-119. |
| [8] | Yao CHEN, Xilu YANG, Xuejia HE. Grazing of three common protozoan on brown tide alga Aureococcus anophagefferens [J]. Journal of Tropical Oceanography, 2018, 37(6): 120-132. |
| [9] | Jiangxing DONG, Wei SHI, Xiaoyu KONG, Shixi CHEN. Characteristic analysis of the complete mitogenome of Plagiopsetta glossa and a possible mechanism for gene rearrangement [J]. Journal of Tropical Oceanography, 2018, 37(1): 1-11. |
|
||