Journal of Tropical Oceanography ›› 2023, Vol. 42 ›› Issue (5): 76-91.doi: 10.11978/2022216CSTR: 32234.14.2022216
• Marine Biology • Previous Articles Next Articles
Distribution of the microbial Carbohydrate-Active enzymes genes in the surface sediment of the Daya Bay, China
SUN Cuici1,2( ), YUE Weizhong3,4, ZHAO Wenjie1,5, WANG Youshao1,2(
), YUE Weizhong3,4, ZHAO Wenjie1,5, WANG Youshao1,2( )
)
- 1. State Key Laboratory of Tropical Oceanography (South China Sea Institute of Oceanology, Chinese Academy of Sciences), Guangzhou 510301, China
2. Daya Bay Marine Biology Research Station (Chinese Academy of Sciences), Shenzhen 518121, China
3. Marine Environmental Center (South China Sea Institute of Oceanology, Chinese Academy of Sciences), Guangzhou 510301, China
4. Yangjiang Offshore Wind Power Laboratory, Yangjiang 529500, China
5. University of Chinese Academy of Sciences, Beijing 100049, China
-
Received:2022-10-10Revised:2022-11-22Online:2023-09-10Published:2023-01-09 -
Supported by:National Natural Science Foundation of China(42073078); National Natural Science Foundation of China(U1901211); Guangdong Basic and Applied Basic Research Foundation(2020A1515011137); National Key Research and Development Program of China(2017FY100707)
Cite this article
SUN Cuici, YUE Weizhong, ZHAO Wenjie, WANG Youshao. Distribution of the microbial Carbohydrate-Active enzymes genes in the surface sediment of the Daya Bay, China[J].Journal of Tropical Oceanography, 2023, 42(5): 76-91.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Physical parameters during the sampling time"
| 站位 | S1 | S5 | S11 |
|---|---|---|---|
| 经度 | 114°37′59.88″E | 114°33′42.12″E | 114°43′0.12″E |
| 纬度 | 22°31′59.88″N | 22°35′35.88″N | 22°46′0.12″N |
| 水深/m | 19.5 | 13.5 | 8.0 |
| 含水率/% | 38.2 | 35.1 | 62.4 |
| 砂土百分比/% | 13.4 | 5.9 | 0 |
| 粉砂土百分比/% | 64.9 | 71.9 | 62.7 |
| 黏土百分比/% | 21.7 | 22.2 | 37.54 |
| 平均粒径/μm | 6.4 | 6.4 | 7.59 |
| 中值粒径/μm | 6.6 | 6.3 | 7.49 |
| 总有机碳(TOC)/(%·g-1) | 0.50±0.02 | 0.47±0.01 | 1.63±0.05 |
| 硫化物含量/(μg· g-1) | 33.41±1.12 | 81.8±2.37 | 218.7±5.49 |
| 总磷(TP)/(mg·kg-1) | 160.5±8.27 | 130.7±5.18 | 142.4±6.76 |
| 总氮(TN)/(mg·kg-1) | 214.5±8.1 | 248.8±9.3 | 441.6±12.8 |
| TOC : TN | 27.3 | 22.0 | 43.1 |
| 水体叶绿素浓度/(μg·L-1) | 1.47 | 1.51 | 1.76 |

Fig. 4
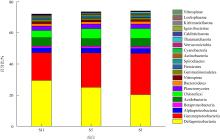
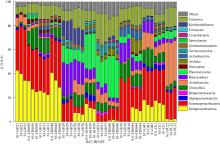
Shannon-diversity (a) and CPCoA analysis of CAZymes families (b) in the different prokaryotic groups (bacteria and Archaea) Aci: Acidobacteria; Act: Actinobacteria; Arc: Archaea; Bac: Bacteroidetes; Chl: Chloroflexi; Cya: Cyanobacteria; Del: δ-proteobacteria; Fir: Firmicutes; Gam: γ-proteobacteria; Gem: Gemmatimonadetes; Kir: Kiritimatiellaeota; Nit: Nitrospirae; Pla: Planctomycetes; Spi: Spirochaetes; Ver: Verrucomicrobia; Kim: Kiritimatiellaeota"
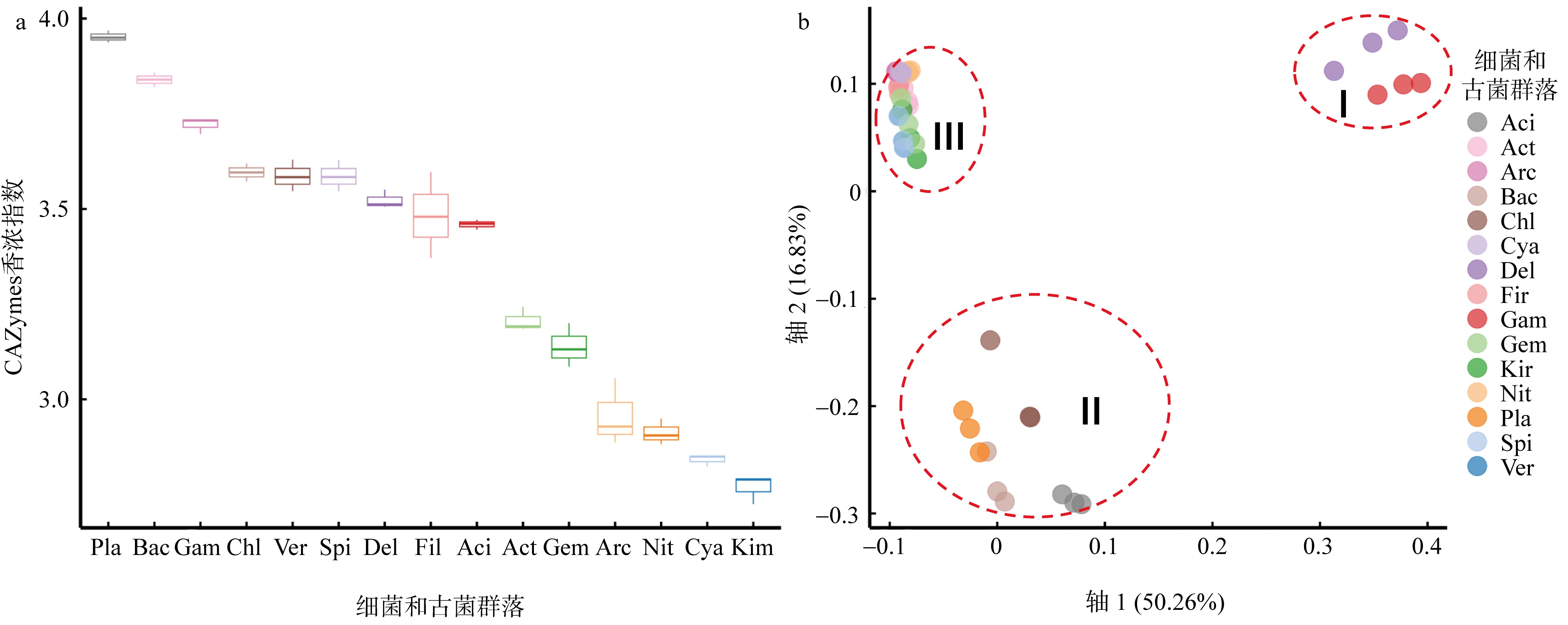
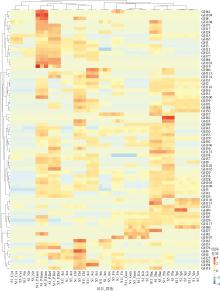
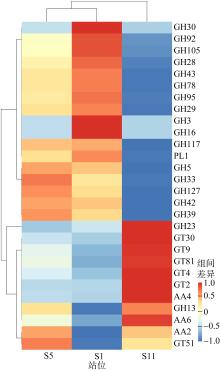
Fig. 5
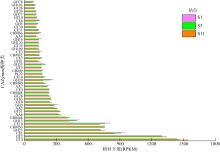
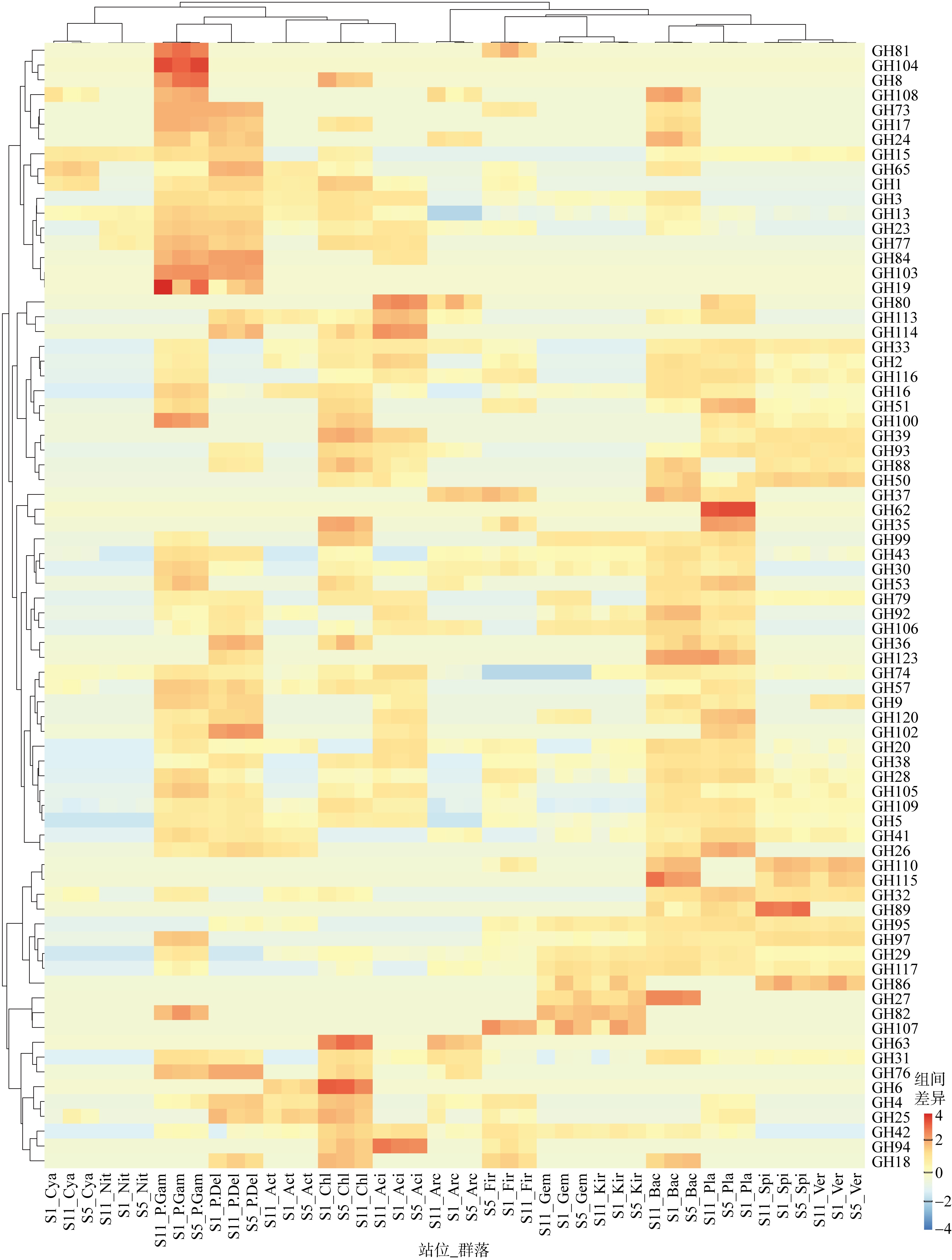
GHs genes distributions in the different groups Aci: Acidobacteria; Act: Actinobacteria; Arc: Archaea; Bac: Bacteroidetes; Chl: Chloroflexi; Cya: Cyanobacteria; P.Del: δ-proteobacteria; Fir: Firmicutes; P.Gam: γ-proteobacteria; Gem: Gemmatimonadetes; Kir: Kiritimatiellaeota; Nit: Nitrospirae; Pla: Planctomycetes; Spi: Spirochaetes; Ver: Verrucomicrobia; Kim: Kiritimatiellaeota"
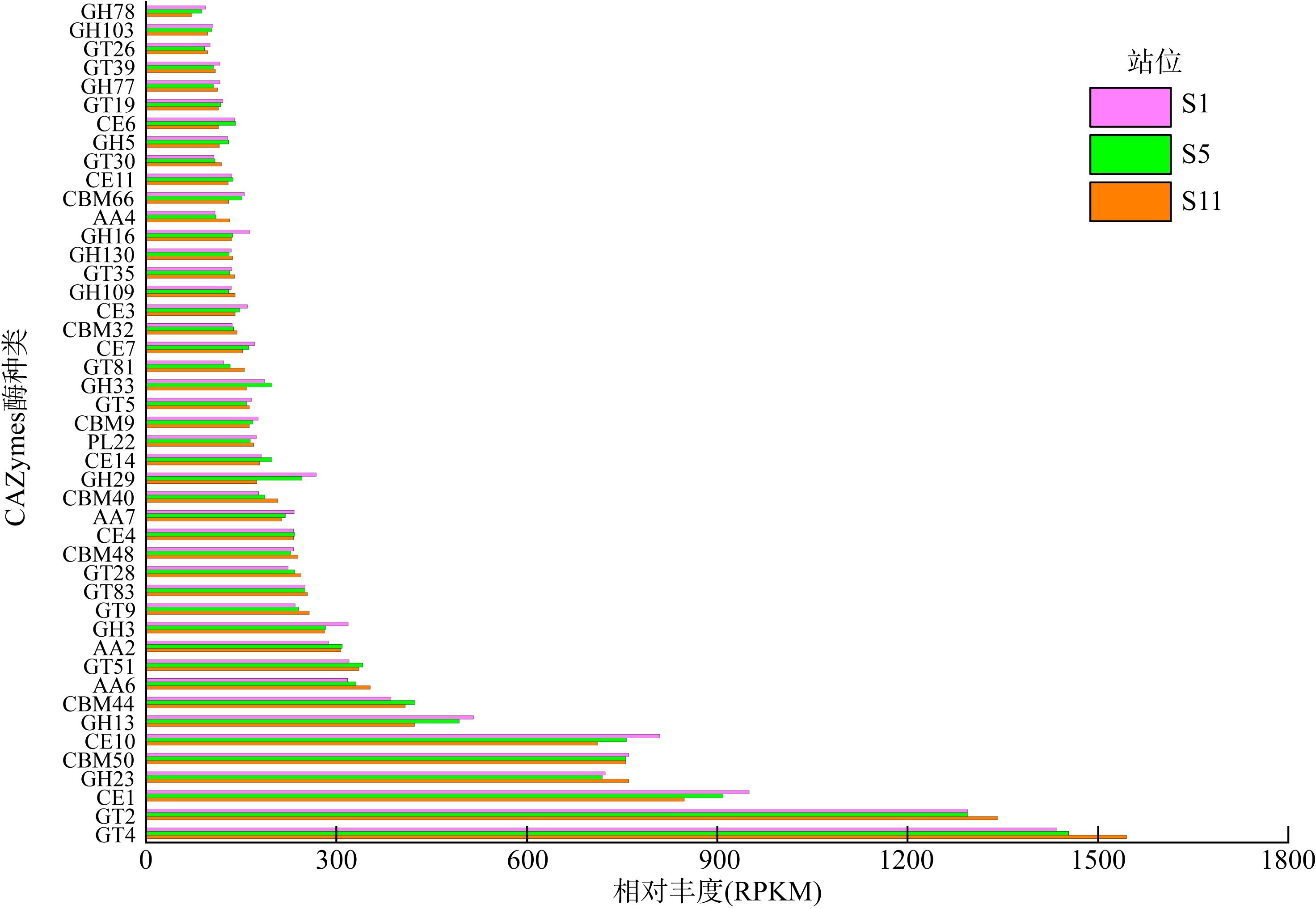
Tab. S1
The major activity of CAZymes"
| 序号 | CAZymes | 名称或主要功能 | ||
|---|---|---|---|---|
| 1 | GH3 | β-葡萄糖苷酶/β-N-乙酰基己糖胺酶 | ||
| 2 | GH4 | α-半乳糖醛酸酶/麦芽糖-6-磷酸葡萄糖苷酶/6-磷酸-β-葡萄糖苷 | ||
| 3 | GH5 | 内葡聚糖酶 | ||
| 4 | GH13 | 异淀粉酶/1,4-α-葡聚糖分支酶/α-葡萄糖苷酶/α淀粉酶 | ||
| 5 | GH15 | 葡聚糖酶/α-海藻糖酶/糖化酶 | ||
| 6 | GH16 | 内切-1,3-β-葡聚糖酶 | ||
| 7 | GH17 | 内切-1,3-β-葡萄糖苷酶 | ||
| 8 | GH18 | 几丁质酶 | ||
| 9 | GH20 | 己糖胺酶 | ||
| 10 | GH23 | 溶菌酶/可溶性溶菌素转糖基酶/膜结合溶菌素转糖基酶 | ||
| 11 | GH29 | α-L-岩藻糖苷酶 | ||
| 12 | GH30 | β-葡萄糖苷酶/葡糖基神经酰胺酶/β-葡萄糖醛酸酶/内切-β-1,6-半乳糖苷酶 | ||
| 13 | GH31 | α-D-木糖苷木糖水解酶/α-葡萄糖苷酶/寡糖4-α-D-葡萄糖基转移酶 | ||
| 14 | GH32 | 果聚糖内切酶 | ||
| 15 | GH33 | 唾液酸酶或神经氨酸酶/反式唾液酸酶/脱水唾液酸酶/2-酮-3-脱氧壬酸水解酶 | ||
| 16 | GH43 | 木聚糖1,4-β-木糖苷酶 | ||
| 17 | GH57 | 1,4-α-葡聚糖分支酶 | ||
| 18 | GH65 | 麦芽糖磷酸化酶/海藻糖磷酸化 | ||
| 19 | GH73 | 溶菌酶 | ||
| 20 | GH77 | 4-α-葡聚糖转移酶 | ||
| 21 | GH78 | α-L-氨苷酶 | ||
| 22 | GH95 | α-L-岩藻糖苷酶2 | ||
| 23 | GH102 | 肽聚糖裂解转糖苷酶 | ||
| 24 | GH103 | 肽聚糖裂解转糖苷酶 | ||
| 25 | GH105 | 不饱和鼠李糖乳糖酰水解酶 | ||
| 26 | GH106 | α-L-鼠李糖苷酶 | ||
| 27 | GH108 | N-乙酰胞壁质酶 | ||
| 28 | GH109 | N-乙酰氨基己糖苷酶 | ||
| 29 | GH127 | β-L-阿拉伯糖苷酶 | ||
| 30 | GH130 | β-1,2-甘露糖磷酸化酶 | ||
| 31 | GH145 | L-鼠李糖-α-1,4-D-葡萄糖醛酸裂解酶 | ||
| 32 | GH149 | β-1,3-葡聚糖磷酸化酶 | ||
| 33 | CE1 | 乙酰木聚糖酯酶/S-甲酰谷胱甘肽水解酶/聚羟基丁酸解聚酶/ /羧酸酯酶 | ||
| 34 | CE2 | 乙酰木聚糖酯酶 | ||
| 35 | CE3 | 乙酰木聚糖酯酶 | ||
| 36 | CE4 | 乙酰木聚糖酯酶/甲壳素脱乙酰酶/壳寡糖脱乙酰酶肽聚糖脱乙酰酶/肽聚糖 N-乙酰胞壁酸脱乙酰酶 | ||
| 37 | CE6 | 乙酰木聚糖酯酶 | ||
| 38 | CE7 | 乙酰木聚糖酯酶 | ||
| 39 | CE8 | 果胶酯酶 | ||
| 40 | CE9 | N-乙酰氨基葡萄糖6-磷酸脱乙酰酶/N-乙酰氨基半乳糖6-磷酸脱乙酰酶 | ||
| 41 | CE10 | 芳香酯酶/羧基酯酶等 | ||
| 42 | CE11 | 尿苷二磷酸-3-O-[3-羟基肉豆蔻酰基]N-乙酰氨基葡萄糖脱乙酰酶/尿苷二磷酸-3-O-[3-羟基肉豆蔻酰]N-乙酰基葡萄糖胺脱乙酰酶/3-羟基酰基-[酰基载体蛋白]脱水酶 | ||
| 43 | CE12 | 果胶乙酰酯酶 | ||
| 44 | CE13 | 果胶乙酰酯酶 | ||
| 45 | CE14 | 二乙酰基壳二糖去乙酰化酶 | ||
| 46 | CE15 | 4-O-甲基葡萄糖醛酸甲酯酶 | ||
| 47 | CE16 | 乙酰酯酶 | ||
| 48 | AA2 | 过氧化氢酶过氧化物酶 | ||
| 49 | AA3 | 葡萄糖-甲醇-胆碱氧化还原酶家族/胆碱脱氢酶 | ||
| 50 | AA4 | 乙醇酸氧化酶/D-乳酸脱氢酶(细胞色素) | ||
| 51 | AA6 | 醌还原酶 | ||
| 52 | AA7 | 葡萄糖氧化酶/壳寡糖氧化酶/纤维低聚糖脱氢酶 | ||
| 53 | AA9 | 依赖铜的裂解多糖单加氧酶 | ||
| 54 | AA12 | 吡咯喹啉醌依赖性氧化还原酶 | ||
| 55 | AA13 | 依赖铜的裂解多糖单加氧酶 | ||
| 56 | PL1 | 果胶裂解酶 | ||
| 57 | PL2 | 果胶裂解酶 | ||
| 58 | PL4 | 鼠李糖半乳糖醛酸内切酶 | ||
| 59 | PL5 | 海藻酸裂解酶 | ||
| 60 | PL6 | 海藻酸裂解酶 | ||
| 61 | PL7 | 海藻酸裂解酶 | ||
| 62 | PL8 | 软骨素裂解酶 | ||
| 63 | PL9 | 果胶裂解酶 | ||
| 64 | PL10 | 果胶裂解酶 | ||
| 65 | PL12 | 硫酸肝素裂解酶 | ||
| 66 | PL14 | 聚(β-甘露糖醛酸)裂解酶/M-特异性海藻酸裂解酶 | ||
| 67 | PL15 | 海藻酸裂解酶/肝素裂解酶/肝素裂解酶 I/肝素-硫酸盐裂解酶/肝素裂解酶 Ⅲ | ||
| 68 | PL17 | 海藻酸裂解酶 | ||
| 69 | PL18 | 海藻酸裂解酶 | ||
| 70 | PL21 | 肝素裂解酶/肝素-硫酸盐裂解酶/阿查兰-硫酸盐裂解酶 | ||
| 71 | PL22 | 低聚半乳糖醛酸裂解酶 | ||
| 72 | PL24 | 石莼多糖裂解酶 | ||
| 73 | CBM6 | 结合纤维素、β-1,4-木聚糖、β-1,13-葡聚糖、β-1, 3-1,4-葡聚糖和β-1,3-葡聚糖 | ||
| 74 | CBM9 | 结合纤维素 | ||
| 75 | CBM11 | 与β-1,4-葡聚糖和β-1,3-1,4-混合连接葡聚糖结合 | ||
| 76 | CBM16 | 结合纤维素和葡甘聚糖 | ||
| 77 | CBM13 | 与纤维素结合 | ||
| 78 | CBM20 | 与淀粉结合 | ||
| 79 | CBM23 | 与甘露聚糖结合 | ||
| 80 | CBM32 | 结合半乳糖、乳糖和N-乙酰-D-乳糖胺 | ||
| 81 | CBM37 | 与木聚糖、几丁质、显微晶质和磷酸纤维素结合 | ||
| 82 | CBM40 | 与GH33结合 | ||
| 83 | CBM41 | 与α-葡聚糖-直链淀粉、支链淀粉、普鲁兰聚糖结合 | ||
| 84 | CBM44 | 结合纤维素和木聚糖 | ||
| 85 | CBM47 | 与岩藻糖结合 | ||
| 86 | CBM48 | 附加到GH13模块 | ||
| 87 | CBM50 | 附着于GH18、GH19、GH23、GH24、GH25和GH73家族的各种酶, 即裂解甲壳素或肽聚糖的酶 | ||
| 88 | CBM57 | 附着于各种糖苷酶 | ||
| 89 | CBM66 | 与果聚糖的果糖苷末端结合, 辅助β-果糖苷外切酶 | ||
| 90 | CBM67 | 与L-鼠李糖结合 | ||
| 91 | GT2 | 纤维素、几丁质、葡聚糖、甘露聚糖、透明质酸合成酶/N-乙酰氨基葡萄糖基转移酶等 | ||
| 92 | GT4 | 蔗糖合成酶/蔗糖-磷酸合成酶/α-葡萄糖基转移酶/脂多糖N-乙酰氨基葡萄糖基转移酶等 | ||
| 93 | GT9 | 脂多糖-乙酰氨基葡萄糖转移酶 | ||
| 94 | GT26 | β-N-乙酰甘露糖苷酰转移酶/β-1,4-葡萄糖基转移酶β-1,4-半乳糖基转移酶 | ||
| 95 | GT28 | 1,2-二酰甘油3-β-半乳糖基转移酶; 1,2-二酰甘油3-β-葡萄糖基转移酶/β-N-乙酰氨基葡萄糖基转移酶/双半乳糖基二酰甘油合成酶 | ||
| 96 | GT30 | α-3-脱氧-D-甘露聚糖-辛酸转移酶 | ||
| 97 | GT35 | 糖原或淀粉磷酸化酶 | ||
| 98 | GT39 | 蛋白α-甘露糖基转移酶 | ||
| 99 | GT51 | 胞壁质聚合酶 | ||
| 100 | GT81 | 葡萄糖基-3-磷酸甘油酸合成酶/甘露糖基 -3-磷酸甘油酸合成酶/ 葡萄糖基-2-甘油酸合成酶/甘露糖基-3-磷酸甘油酸合成酶/3-磷酸甘油酸α-甘露糖基转移酶 | ||
| 101 | GT83 | 4-氨基-4-脱氧-β-L-阿拉伯糖基转移酶/半乳糖醛酸基转移酶 | ||
| [1] |
牟新悦, 陈敏, 张琨, 等, 2017. 夏季大亚湾悬浮颗粒有机物碳、氮同位素组成及其物源指示[J]. 海洋学报, 39(2): 39-52.
|
|
|
|
| [2] |
中华人民共和国国家质量监督检验检疫总局和中国国家标准化管理委员会, 2007. 海洋监测规范(GB17378. 5—2007)[S]. 北京: 中国标准出版社 (in Chinese).
|
| [3] |
doi: 10.1007/s00248-016-0911-9 pmid: 28070679 |
| [4] |
doi: 10.1016/j.earscirev.2013.02.008 |
| [5] |
doi: 10.1146/marine.2021.13.issue-1 |
| [6] |
doi: 10.1002/lno.v66.9 |
| [7] |
doi: 10.1186/s40168-021-01063-4 pmid: 33975640 |
| [8] |
doi: 10.1038/s41396-021-00928-8 pmid: 33649555 |
| [9] |
doi: 10.1111/1462-2920.15550 pmid: 33973326 |
| [10] |
doi: 10.1093/bioinformatics/btu170 pmid: 24695404 |
| [11] |
pmid: 21141030 |
| [12] |
doi: 10.1371/journal.pone.0028742 |
| [13] |
doi: 10.1038/s41467-022-31433-x pmid: 35790765 |
| [14] |
doi: 10.1186/s40168-020-00836-7 pmid: 32482164 |
| [15] |
doi: 10.1016/j.ymeth.2013.06.027 pmid: 23816787 |
| [16] |
|
| [17] |
doi: 10.1111/emi4.2014.6.issue-6 |
| [18] |
doi: 10.1186/s40793-021-00385-y pmid: 34404489 |
| [19] |
doi: 10.1016/j.cell.2021.12.036 |
| [20] |
doi: 10.1007/s00253-009-2331-y pmid: 19908036 |
| [21] |
doi: 10.1038/s41396-020-0588-4 pmid: 31988474 |
| [22] |
|
| [23] |
doi: 10.1038/s41396-018-0252-4 pmid: 30116038 |
| [24] |
doi: 10.1111/1462-2920.13142 pmid: 26626228 |
| [25] |
doi: 10.1038/nature12033 |
| [26] |
doi: 10.1038/s41467-021-22765-1 pmid: 33927199 |
| [27] |
doi: 10.1038/ismej.2013.174 pmid: 24108328 |
| [28] |
doi: 10.1007/s00284-016-1089-6 pmid: 27363425 |
| [29] |
doi: 10.1038/s41564-017-0047-9 pmid: 29062087 |
| [30] |
doi: 10.1038/s41564-020-0720-2 pmid: 32451471 |
| [31] |
doi: 10.3389/fmicb.2019.02475 |
| [32] |
doi: 10.1126/science.aan4834 pmid: 28839072 |
| [33] |
doi: 10.1038/ismej.2016.84 pmid: 27300277 |
| [34] |
doi: 10.1038/ismej.2016.24 pmid: 26943623 |
| [35] |
doi: 10.1126/science.1218344 pmid: 22556258 |
| [36] |
doi: 10.7554/eLife.11888 |
| [37] |
doi: 10.3389/fmicb.2019.00253 |
| [38] |
doi: 10.1038/s41467-021-21009-6 |
| [39] |
doi: 10.1128/AEM.00055-15 |
| [40] |
doi: 10.1016/j.envpol.2021.117267 |
| [41] |
doi: 10.1038/ismej.2014.225 pmid: 25478683 |
| [42] |
|
| [43] |
doi: 10.1093/nar/gky418 |
| [44] |
doi: 10.1126/sciadv.aaz4354 |
| [45] |
doi: 10.1111/1462-2920.15650 pmid: 34196089 |
| [1] | CHEN Jie, BIAN Cheng, JIANG Changbo, YAO Zhen, JIANG Chao, LIANG Hai. Advances in the characterization of bioclastic sediment motion [J]. Journal of Tropical Oceanography, 2024, 43(4): 33-41. |
| [2] | MO Danyang, NING Zhiming, YANG Bin, XIA Ronglin, LIU Zhijin. Response of dissimilatory nitrate reduction processes in coral reef sediments of the Weizhou island to temperature changes [J]. Journal of Tropical Oceanography, 2024, 43(4): 137-143. |
| [3] | XI Chen, LIN Zongxuan, SA Rula, DENG Xi, LIU Qiang, NI Liang, LUO Laicai, MA Teng, XIE Zhijie, CHEN Siruo, CHEN Songze. Analysis of water environmental changes and influencing factors in the southwestern waters of the Daya Bay based on continuous monitoring data from dual buoys [J]. Journal of Tropical Oceanography, 2024, 43(4): 153-164. |
| [4] | GAO Jie, YU Kefu, XU Shendong, HUANG Xueyong, CHEN Biao, WANG Yonggang. Content and source analysis of organic carbon in the outer slope sediments of the Yongle Atoll, Xisha Islands [J]. Journal of Tropical Oceanography, 2024, 43(3): 131-145. |
| [5] | XING Nannan, REN Runxin, TANG Zhenzhou, LUO Zhihong, XIA Chenxi, LIU Yonghong, PENG Liang, CHEN Xianqiang. Study on the secondary metabolites of fungus Aspergillus sp. GXIMD02003 derived from marine sediment in the Weizhou island [J]. Journal of Tropical Oceanography, 2023, 42(5): 154-160. |
| [6] | YANG Xialing, LI Shushi, XU Shanshan, YU Chongxi, PAN Jie. Variations in water and sediments of the Nanliu River flowing into the sea under the influence of extreme weather in the past 60 years [J]. Journal of Tropical Oceanography, 2023, 42(4): 91-103. |
| [7] | SONG Xingyu, LIN Yajun, ZHANG Liangkui, XIANG Chenhui, HUANG Yadong, ZHENG Chuanyang. Distribution characteristics and influencing factors of meso- and micro-zooplankton communities in the offshore waters of the Guangdong-Hong Kong-Macao Greater Bay Area* [J]. Journal of Tropical Oceanography, 2023, 42(3): 136-148. |
| [8] | ZHANG Min, WU Hangxing, LU Yibin, LU Diwen, MI Jie, ZHU Donglin, CHEN Bo. Impact of the coastal reclamations on topography evolution in the Qinzhou Bay, Guangxi [J]. Journal of Tropical Oceanography, 2023, 42(2): 124-131. |
| [9] | JIANG Xun, WU Wen, SONG Dehai. Identification and quantitative analysis of key controlling factors of water quality response to human activities in the Daya Bay, China [J]. Journal of Tropical Oceanography, 2023, 42(1): 182-191. |
| [10] | YANG Hongqiang, TAN Fei, XU Huilong, ZHANG Xiyang, SHI Qi, TAO Shichen. Reconstruction of the tropical cyclones activity in the Nansha Islands since the Little Ice Age from the atoll lagoon sediments [J]. Journal of Tropical Oceanography, 2022, 41(6): 171-182. |
| [11] | CHEN Jingfu, ZHONG Yu, WANG Lei, GUO Yupei, QIU Dajun. Noctiluca scintillans effects on eukaryotic plankton community structure using Environmental DNA analysis in Daya Bay* [J]. Journal of Tropical Oceanography, 2022, 41(5): 121-132. |
| [12] | HUANG Zuming, ZHOU Xiaoyan, DAI Zhijun, CHE Zhiwei. Analysis of the variations of suspended sediment concentration in vertical profile near the bed of Aegiceras corniculatum tidal flat [J]. Journal of Tropical Oceanography, 2022, 41(4): 38-50. |
| [13] | LI Weihua, LI Jiufa, ZHANG Wenxiang. Research progress in the continuous measurement technology of suspended sediment concentration [J]. Journal of Tropical Oceanography, 2022, 41(4): 20-30. |
| [14] | LI Zhen, LI Yunkai, LIU Yonghu, CHENG Qian, ZHANG Shuo. Release potential of different forms of nitrogen extracted by chemical leaching in the surface sediments of the Bohai Sea [J]. Journal of Tropical Oceanography, 2022, 41(4): 163-171. |
| [15] | WANG Zhaohui, ZHANG Yuning, WANG Wenting, XIE Changliang, CHEN Jiazhuo, ZHENG Hu, WANG Junxing. Distribution of dinoflagellate cysts in surface sediments on the Dongshan Bay, Fujian province, China [J]. Journal of Tropical Oceanography, 2022, 41(4): 154-162. |
|
||