Journal of Tropical Oceanography ›› 2020, Vol. 39 ›› Issue (4): 42-49.doi: 10.11978/2019115CSTR: 32234.14.2019115
• Marine Biology • Previous Articles Next Articles
Genetic polymorphism of the Brushtooth lizardfish Saurida undosquamis based on mitochondrial D-loop sequences
Min LI1,2,3( ), Xiaolan KONG1, Youwei XU1, Zuozhi CHEN1,2,3(
), Xiaolan KONG1, Youwei XU1, Zuozhi CHEN1,2,3( )
)
- 1. Key Laboratory of Open-Sea Fishery Development, Ministry of Agriculture and Rural Affairs, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou 510300, China
2. Guangdong Provincial Key Laboratory of Fishery Ecology and Environment, South China Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou 510300, China
3. Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou), Guangzhou 511458, China
-
Received:2018-11-13Revised:2020-01-11Online:2020-07-20Published:2020-07-27 -
Contact:Zuozhi CHEN E-mail:limin@scsfri.ac.cn;zzchen2000@163.com -
Supported by:Foundation item: Natural Science Foundation of Guangdong Province(2014A030310177);Financial Fund of the Ministry of Agriculture and Rural Affairs(NFZX2018);Special Fund for Promoting Marine Economic Development of Guangdong Province(GDME-2018E004);Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS(2019TS13)
CLC Number:
- P735
Cite this article
Min LI, Xiaolan KONG, Youwei XU, Zuozhi CHEN. Genetic polymorphism of the Brushtooth lizardfish Saurida undosquamis based on mitochondrial D-loop sequences[J].Journal of Tropical Oceanography, 2020, 39(4): 42-49.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
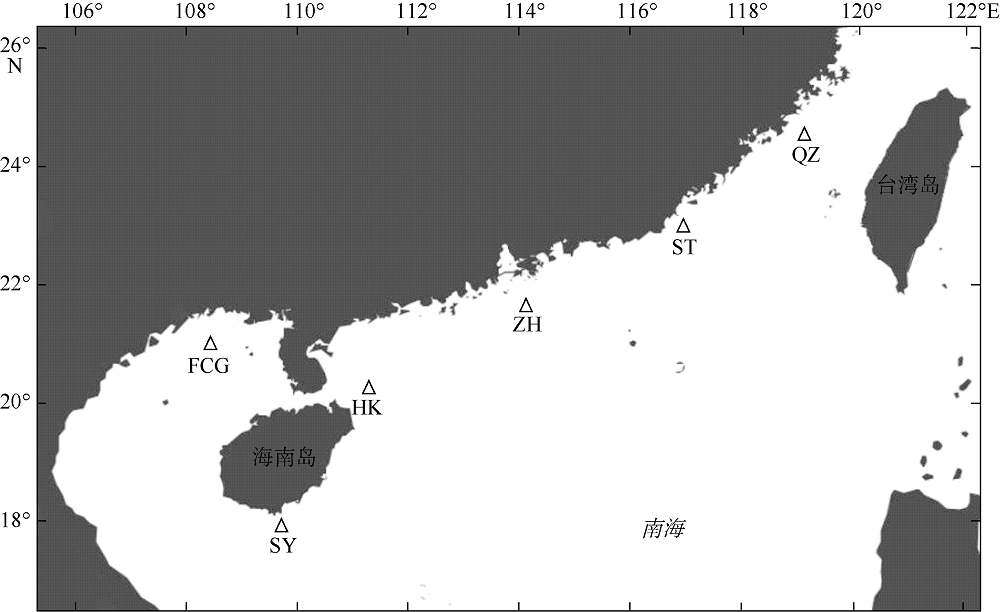
Tab. 1
Specimen information of S. undosquamis and genetic diversity parameters based on D-loop sequences"
| 地理群体(缩写) | 采样点位置 | 样本量/个 | 单倍型数量/个 | 单倍型多样性(h±SD) | 核苷酸多样性(π±SD) |
|---|---|---|---|---|---|
| 防城港(FCG) | 108°30'E, 21°00'N | 21 | 19 | 0.9905 ± 0.0178 | 0.0110 ± 0.0058 |
| 三亚(SY) | 109°46'E, 17°58'N | 22 | 21 | 0.9957 ± 0.0153 | 0.0148 ± 0.0077 |
| 海口(HK) | 111°18'E, 20°18'N | 22 | 21 | 0.9957 ± 0.0153 | 0.0135 ± 0.0071 |
| 珠海(ZH) | 114°05'E, 21°41'N | 21 | 20 | 0.9952 ± 0.0165 | 0.0142 ± 0.0074 |
| 汕头(ST) | 116°55'E, 23°00'N | 22 | 19 | 0.9827 ± 0.0208 | 0.0129 ± 0.0068 |
| 泉州(QZ) | 119°02'E, 24°36'N | 21 | 21 | 1.0000 ± 0.0147 | 0.0128 ± 0.0068 |
| 总计 | / | 129 | 101 | 0.9873 ± 0.0048 | 0.0132 ± 0.0067 |
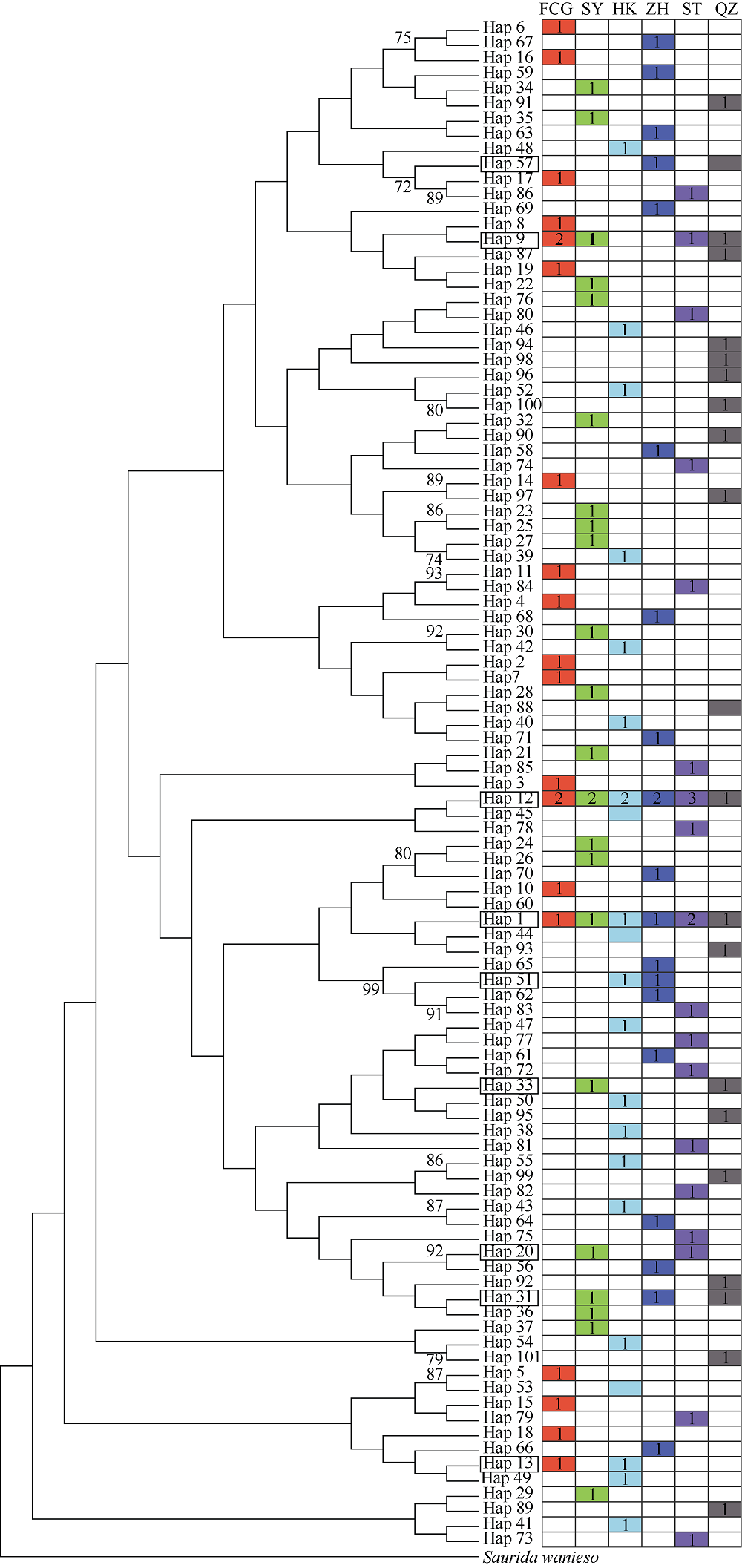
Fig. 2
Topology of NJ tree for S. undosquamis D-loop sequence haplotypes, and distribution of haplotypes among geographic populations. Bootstrap values of >70% are shown at nodes (percentage). Saurida wanieso is used as outgroup. The shared haplotypes are framed. The colors represent the corresponding geographic populations, and the numbers represent the haplotype frequencies"
Tab. 3
Pairwise FST (below diagonal) and P values (above diagonal) among geographic populations of S. undosquamis based on D-loop sequences"
| FCG | SY | HK | ZH | ST | QZ | |
|---|---|---|---|---|---|---|
| FCG | 0.0773 | 0.0707 | 0.0202 | 0.1060 | 0.1605 | |
| SY | 0.0269 | 0.9300 | 0.6829 | 0.8408 | 0.7405 | |
| HK | -0.0267 | -0.0211 | 0.7952 | 0.9263 | 0.8341 | |
| ZH | 0.0478 | -0.0126 | -0.0155 | 0.9421 | 0.2658 | |
| ST | -0.0506 | -0.0176 | -0.0204 | -0.0238 | 0.3424 | |
| QZ | 0.0164 | -0.0155 | -0.0178 | 0.0071 | 0.0017 |
| [1] | 蔡研聪, 徐姗楠, 陈作志, 等, 2018. 南海北部近海渔业资源群落结构及其多样性现状[J]. 南方水产科学, 14(2):10-18. |
| CAI YANCONG, XU SHANNAN, CHEN ZUOZHI, et al, 2018. Current status of community structure and diversity of fishery resources in offshore northern South China Sea[J]. South China Fisheries Science, 14(2):10-18 (in Chinese with English abstract). | |
| [2] | 陈再超, 刘继兴, 1982. 南海经济鱼类[M]. 广州: 广东科学与技术出版社: 184-188(in Chinese). |
| [3] | 陈作志, 邱永松, 徐姗楠, 等, 2012. 北部湾花斑蛇鲻生物学特征的演化[J]. 中国水产科学, 19(2):321-328. |
| CHEN ZUOZHI, QIU YONGSONG, XU SHANNAN, et al, 2012. Evolution of biological characteristics of Saurida undosquamis (Richardson) in the Beibu Gulf, South China Sea[J]. Journal of Fishery Sciences of China, 19(2):321-328 (in Chinese with English abstract). | |
| [4] | 郜星晨, 章群, 薛丹, 等, 2016. 基于线粒体控制区部分序列的南海大斑石鲈遗传多样性分析[J]. 海洋科学, 40(7):41-45. |
| GAO XINGCHEN, ZHANG QUN, XUE DAN, et al, 2016. Genetic diversity of Pomadasys maculatus in coastal waters of South China Sea via mtDNA control region partial sequences[J]. Marine Sciences, 40(7):41-45 (in Chinese with English abstract). | |
| [5] | 黄小林, 李文俊, 林黑着, 等, 2018. 基于线粒体DNA D-loop序列的黄斑篮子鱼群体遗传多样性分析[J]. 热带海洋学报, 37(4):45-51. |
| HUANG XIAOLIN, LI WENJUN, LIN HEIZHAO, et al, 2018. Genetic variations among Siganus oramin populations in coastal waters of southeast China based on mtDNA control region sequences[J]. Journal of Tropical Oceanography, 37(4):45-51 (in Chinese with English abstract). | |
| [6] | 黄梓荣, 2002. 休渔对南海北部多齿蛇鲻资源的影响[J]. 湛江海洋大学学报, 22(6):26-31. |
| HUANG ZIRONG, 2002. Effect of Close Season on Bloch et Schneider (S. tumbil) Stock in the North of South China Sea[J]. Journal of Zhanjiang Ocean University, 22(6):26-31 (in Chinese with English abstract). | |
| [7] | 江艳娥, 许友伟, 范江涛, 等, 2019. 南海北部陆架水域多齿蛇鲻与花斑蛇鲻的年龄与生长[J]. 中国水产科学, 26(1):82-90. |
| JIANG YAN’E, XU YOUWEI, FAN JIANGTAO, et al, 2019. Age and growth of Saurida tumbil and Saurida undosquamis in the continental shelf of the northern South China Sea[J]. Journal of Fishery Sciences of China, 26(1):82-90 (in Chinese with English abstract). | |
| [8] | 梁述章, 宋炜, 马春艳, 等, 2019. 基于线粒体控制区的中国近海棘头梅童鱼群体遗传结构研究[J]. 海洋渔业, 41(2):138-148. |
| LIANG SHUZHANG, SONG WEI, MA CHUNYAN, et al, 2019. Genetic structure of Collichthys lucidus populations from China coastal waters based on mt DNA control region[J]. Marine Fisheries, 41(2):138-148 (in Chinese with English abstract). | |
| [9] | 吕金磊, 章群, 杨喜书, 等, 2017. 基于线粒体控制区的中国南海海域卵形鲳鲹遗传多样性[J]. 海洋渔业, 39(3):241-248. |
| LV JINLEI, ZHANG QUN, YANG XISHU, et al, 2017. Genetic diversity of Trachinotus ovatus in the South China Sea inferred from Mitochondrial DNA control region sequences[J]. Marine Fisheries, 39(3):241-248 (in Chinese with English abstract). | |
| [10] | 舒黎明, 邱永松, 2004a. 南海北部花斑蛇鲻生长死亡参数估计及开捕规格[J]. 湛江海洋大学学报, 24(3):29-35. |
| SHU LIMING, QIU YONGSONG, 2004a. Estimate for growth, mortality parameters and first capture specification suggestion of Saurida undosquamis (Richardson) in Northern South China Sea[J]. Journal of Zhanjiang Ocean University, 24(3):29-35 (in Chinese with English abstract). | |
| [11] | 舒黎明, 邱永松, 2004b. 南海北部多齿蛇鲻生物学分析[J]. 中国水产科学, 11(2):154-158. |
| SHU LIMING, QIU YONGSONG, 2004b. Biology analysis of Saurida tumbil in northern South China Sea[J]. Journal of Fishery Sciences of China, 11(2):154-158 (in Chinese with English abstract). | |
| [12] | 苏纪兰, 袁业立, 2005. 中国近海水文[M]. 北京: 海洋出版社: 1-367(in Chinese). |
| [13] | 孙冬芳, 董丽娜, 李永振, 等, 2010. 南海北部海域多齿蛇鲻的种群分析[J]. 水产学报, 34(9):1387-1394. |
| SUN DONGFANG DONG LI’NA LI YONGZHEN, et al, 2010. Population analysis of Saurida tumbil in the Northern South China Sea[J]. Journal of Fisheries of China, 2010,34(9):1387-1394 (in Chinese with English abstract). | |
| [14] | 孙典荣, 林昭进, 2004. 北部湾主要经济鱼类资源变动分析及保护对策探讨[J]. 热带海洋学报, 23(2):62-68. |
| SUN DIANRONG, LIN ZHAOJIN, 2004. Variations of major commercial fish stocks and strategies for fishery management in beibu gulf[J]. Journal of Tropical Oceanography, 23(2):62-68 (in Chinese with English abstract). | |
| [15] | 熊丹, 李敏, 陈作志, 等, 2015. 南海短尾大眼鲷的种群遗传结构分析[J]. 南方水产科学, 11(2):27-34. |
| XIONG DAN, LI MIN, CHEN ZUOZHI, et al, 2015. Genetic structure of Priacanthus macracanthus population from the South China Sea[J]. South China Fisheries Science, 11(2):27-34 (in Chinese with English abstract). | |
| [16] | 许友伟, 陈作志, 范江涛, 等, 2015. 南沙西南陆架海域底拖网渔获物组成及生物多样性[J]. 南方水产科学, 11(5):76-81. |
| XU YOUWEI, CHEN ZUOZHI, FAN JIANGTAO, et al, 2015. Composition and biodiversity of catches by bottom trawl fishery in southwestern continent shelf of Nansha area[J]. South China Fisheries Science, 2015,11(5):76-81 (in Chinese with English abstract). | |
| [17] | 杨炳忠, 杨吝, 谭永光, 等, 2017. 南海区2种桁杆虾拖网渔获组成调查与分析[J]. 南方水产科学, 13(6):115-122. |
| YANG BINGZHONG, YANG LIN, TAN YONGGUANG, et al, 2017. Analysis of catch composition of two shrimp beam trawls in South China Sea[J]. South China Fisheries Science, 13(6):115-122 (in Chinese with English abstract). | |
| [18] | 杨喜书, 章群, 薛丹, 等, 2018. 中国近海细鳞鯻线粒体控制区的遗传多样性[J]. 生态学报, 38(5):1852-1859. |
| YANG XISHU, ZHANG QUN, XUE DAN, et al, 2018. Genetic analysis of Terapon jarbua in coastal waters of China by using mitochondrial DNA control region sequencing[J]. Acta Ecologica Sinica, 38(5):1852-1859 (in Chinese with English abstract). | |
| [19] | 张俊, 陈国宝, 陈作志, 等, 2015. 南沙南部陆架海域渔业资源声学评估[J]. 南方水产科学, 11(5):1-10. |
| ZHANG JUN, CHEN GUOBAO, CHEN ZUOZHI, et al, 2015. Acoustic estimation of fishery resources in southern continental shelf of Nansha area[J]. South China Fisheries Science, 11(5):1-10 (in Chinese with English abstract). | |
| [20] | ALZOHAIRY A M, 2011. BioEdit: an important software for molecular biology[J]. GERF Bulletin of Biosciences, 2(1):60-61. |
| [21] | CADRIN S X, KERR LA, MARIAN S, 2014. Chapter One-Stock identification methods: an overview[M]//CADRIN S X, KERR LA, MARIAN S. Stock Identification Methods. 2nd ed. San Diego: Elsevier Inc.: 1-5. |
| [22] | CHHANDAPRAJNADARSINI E M, ROUL S K, SWAIN S, et al, 2018. Biometric analysis of brushtooth lizard fish Saurida undosquamis (Richardson, 1848) from Mumbai waters[J]. Journal of Entomology and Zoology Studies, 6(2):1165-1171. |
| [23] | COATES D J, BYRNE M, MORITZ C, 2018. Genetic diversity and conservation units: dealing with the species-population continuum in the age of genomics[J]. Frontiers in Ecology and Evolution, 6:165. |
| [24] |
EXCOFFIER L, LISCHER H E L, 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows[J]. Molecular Ecology Resources, 10(3):564-567.
pmid: 21565059 |
| [25] | FRANKHAM R, BALLOU J D, BRISCOE D A, 2002. Introduction to conservation genetics[M]. Cambridge: Cambridge University Press: 78-104. |
| [26] | FROESE R, PAULY D, (20190801)[20190925] FishBase [DB/OL]. https://www.fishbase.in/summary/Saurida-undosquamis.html |
| [27] | GRANT W S, BOWEN B W, 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation[J]. Journal of Heredity, 89(5):415-426. |
| [28] |
HEWITT G M, 2000. The genetic legacy of the Quaternary ice ages[J]. Nature, 405(6789):907-913.
doi: 10.1038/35016000 pmid: 10879524 |
| [29] |
KUMAR S, STECHER G, TAMURA K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets[J]. Molecular Biology and Evolution, 33(7):1870-1874.
doi: 10.1093/molbev/msw054 pmid: 27004904 |
| [30] |
LAIKRE L, PALM S, RYMAN N, 2005. Genetic population structure of fishes: Implications for coastal zone management[J]. Ambio, 34(2):111-119.
pmid: 15865307 |
| [31] | MALI K S, KUMAR M V, FAREJIYA M K, et al, 2017. Reproductive biology of Saurida tumbil (Bloch 1795) and Saurida undosquamis (Richardson 1848) inhabiting Northwest coast of India[J]. International Journal of Pure & Applied Bioscience, 5(6):957-964. |
| [32] | NAJMUDEEN T M, SEETHA P K, ZACHARIA P U, 2019. Stock dynamics of the brushtooth lizardfish Saurida undosquamis (Richardson, 1848) from a tropical multispecies fishery in the southeastern Arabian Sea[J]. Aquatic Living Resources, 32(5):9. |
| [33] |
PALSBØLL P J, BÉRUBÉ M, ALLENDORF F W, 2007. Identification of management units using population genetic data[J]. Trends in Ecology & Evolution, 22(1):11-16.
pmid: 16982114 |
| [34] | PALUMBI S R, 1994. Genetic divergence, reproductive isolation, and marine speciation[J]. Annual Review of Ecology and Systematics, 25:547-572. |
| [35] |
PINSKY M L, PALUMBI S R, 2014. Meta-analysis reveals lower genetic diversity in overfished populations[J]. Molecular Ecology, 23(1):29-39.
doi: 10.1111/mec.12509 pmid: 24372754 |
| [36] | REISS H, HOARAU G, DICKEY-COLLAS M, et al, 2009. Genetic population structure of marine fish: mismatch between biological and fisheries management units[J]. Fish and Fisheries, 10(4):361-395. |
| [37] |
ROZAS J, FERRER-MATA A, SÁNCHEZ-DELBARRIO J C, et al, 2017. DnaSP 6: DNA sequence polymorphism analysis of large data sets[J]. Molecular Biology and Evolution, 34(12):3299-3302.
doi: 10.1093/molbev/msx248 pmid: 29029172 |
| [1] | REN Huimin, ZHANG Heng, XU Shasha, LI Shengfa, LI Jiansheng, LI Zhihong, HE Lijun. Population genetic structure and historical dynamics of Trachurus japonicus in the China seas based on mitochondrial control region [J]. Journal of Tropical Oceanography, 2021, 40(5): 36-44. |
| [2] | ZOU Congcong, WANG Lijuan, WU Zhihao, YOU Feng. Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences* [J]. Journal of Tropical Oceanography, 2021, 40(5): 25-35. |
| [3] | HUANG Jing, OU Zhekui, LIU Wenguang, HE Maoxian. Genetic Structure and Diversity Analysis of Three Natural Populations of Tectus pyramis Based on Specific Locus Amplified Fragment Sequencing* [J]. Journal of Tropical Oceanography, 2020, 39(5): 1-18. |
| [4] | HUANG Jing, PAN Xiaolan, XU Meng, LIU Wenguang, ZHANG Hua, HE Maoxian. Morphological and SNP markers for analysis of genetic structure of hybrid progeny and their parental populations of Pinctada fucata martensii [J]. Journal of Tropical Oceanography, 2019, 38(6): 80-89. |
| [5] | Xiaolin HUANG, Wenjun LI, Heizhao LIN, Yukai YANG, Tao LI, Wei YU, Zhong HUANG. Genetic variations among Siganus oramin populations in coastal waters of southeast China based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2018, 37(4): 45-51. |
| [6] | Zhiying ZHAO, Liyun LIANG, Lirong BAI. Analysis of genetic diversity among three wild populations of Penaeus monodon using microsatellite marker [J]. Journal of Tropical Oceanography, 2018, 37(3): 65-72. |
| [7] | WENG Zhaohong, XIE Yangjie, XIAO Zhiqun, WANG Yilei. Analysis of genetic diversity in several wild and hatchery populations of Crassostrea angulata from south Fujian and south Guangdong [J]. Journal of Tropical Oceanography, 2016, 35(3): 94-98. |
| [8] | SHI Xiao-feng1, 2, SU Yong-quan1, WANG Wen-cheng1, WANG Jun1. Population genetic structure of three stocks of Acanthopagrus schlegelii based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2015, 34(1): 56-63. |
| [9] | LIU Li, ZHAO Jie, GUO Yu-song, LIU Chu-wu. Study on the AFLP primer selection and genetic diversities of 4 Snappers [J]. Journal of Tropical Oceanography, 2012, 31(4): 112-116. |
| [10] | LI Li-hao,YU Da-hui,HUANG Gui-ju,DU Bo,FU Yun,TONG Xin,GUO Yi-hui,YE Wei. Comparison of genetic diversity among stocks of Oreochromis niloticus, O. aureus and red tilapia based on microsatellite DNA [J]. Journal of Tropical Oceanography, 2012, 31(2): 102-109. |
| [11] | JI Lei,OU You-jun,LI Jia-er. Genetic polymorphism of three cultured populations of golden pompano Trachinotus ovatus as revealed by microsatellites [J]. Journal of Tropical Oceanography, 2011, 30(3): 62-68. |
| [12] | GUO Yu-song,WANG Zhong-duo,LIU Chu-wu,CHEN Zhi-ming,LIU Yun. Isolation and genetic diversity analysis of microsatellites from nine species of familiar snappers [J]. Journal of Tropical Oceanography, 2010, 29(3): 82-86. |
|
||