Journal of Tropical Oceanography ›› 2018, Vol. 37 ›› Issue (3): 65-72.doi: 10.11978/2017084CSTR: 32234.14.2017084
• Orginal Article • Previous Articles Next Articles
Analysis of genetic diversity among three wild populations of Penaeus monodon using microsatellite marker
Zhiying ZHAO1,2( ), Liyun LIANG2, Lirong BAI1,2
), Liyun LIANG2, Lirong BAI1,2
- 1. Hainan Academy of Ocean and Fisheries Sciences, Haikou 571126 China
2. Hainan Key Laboratory of Tropical Marine Culture Technology, Qionghai 571429, China
-
Received:2017-07-30Revised:2017-10-17Online:2018-06-10Published:2018-05-03 -
Supported by:Application Technology R & D and Demonstration Project of Hainan Province (ZDXM2015025)
Cite this article
Zhiying ZHAO, Liyun LIANG, Lirong BAI. Analysis of genetic diversity among three wild populations of Penaeus monodon using microsatellite marker[J].Journal of Tropical Oceanography, 2018, 37(3): 65-72.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
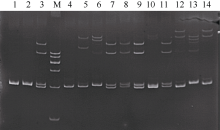
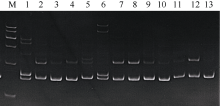
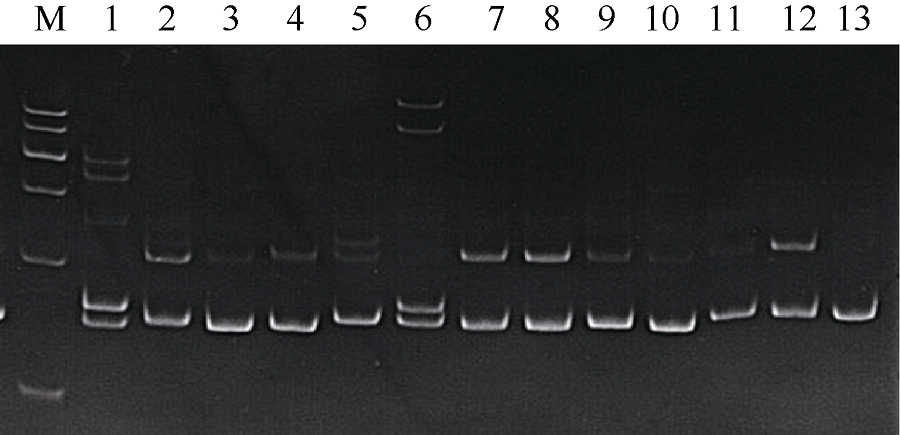
Tab. 1
Fourteen pairs of effective microsatellites primers sequence and their annealing temperature"
| 微卫星位点 | GenBank序列号 | 引物序列(5′-3′) | 退火温度/℃ |
|---|---|---|---|
| DTLPM218 | AY 189727 | F: ATTCCGCAATATATCGGTTTCC | 57 |
| R: AATGTTTCCATTTCATGCTTCG | |||
| DTLPM303 | AY 188984 | F: TGCCTTGTATTTTGACGATCAG | 56 |
| R: TTGGAGTAGCAACAGCGGTA | |||
| DTLPM402 | AY 188987 | F: CCACTCTAACTCCGCCAGTC | 54 |
| R: TCCCTACCCCACTATCATCG | |||
| DTLPM 120 | AY188978 | F: TTATCCGTATAGCCGCGTTATC | 52 |
| R: TTACAGGACCTGCATTTGTGTC | |||
| DTLPM 104 | AY188974 | F: AGGACCTGCATTTGTGTCG | 52 |
| R: ATGGCGAGACAAGGTTCG | |||
| BC52 | AF097631 | F: AAGCAGATAGACCGTCAG | 52 |
| R: AATCCTGCAGAGCAATAC | |||
| TUZXPM49 | AF077568 | F: ATCTGACAGGGCACCATAC | 54 |
| R: AGTCGAGTCTTGAATAAGCG | |||
| PM138 | P1-AY500853 | F: ACGGAGTGGGTAGAGACATA | 56 |
| R: ACAAGCGAAGTGAAGAGG | |||
| PM205 | AY500854 | F: AGGAATGATGGGAGGGAAAG | 56 |
| R: AAGCTCAGGCAAGCGTGTAT | |||
| PM528 | AY500855 | F: GTGTTATTTTCCACGGGTGC | 56 |
| R: GCTGCAGGAAGTGTAGGGAG | |||
| PM1713 | AY500858 | F: GTTGCGACGGGTTGATTC | 54 |
| R: TTTATGGCTATGGCTGACAC | |||
| PM2345 | AY500860 | F: GATATTTCAAGGAATGCTCG | 54 |
| R: TAATTCGTGCCTTACCTCAT | |||
| PM3852 | AY500862 | F: TAATGGGCGTAAGTCTTCGG | 56 |
| R: TGAAAGGAGTCGGGATATGC | |||
| PM4793 | AY500867 | F: CTTCTAGCGCCATTTCAAGG | 56 |
| R: TCCTTCCAGTGTTCGGAGTT |
Tab. 2
Allelic frequency of 14 microsatellite loci in three populations of P. monodon"
| 位点 | 等位基因 | 群体 | 位点 | 等位基因 | 群体 | ||||
|---|---|---|---|---|---|---|---|---|---|
| 琼海 | 三亚 | 湛江 | 琼海 | 三亚 | 湛江 | ||||
| PM1384 | A | 0.500 | 0.4667 | 0.4167 | PM1383 | A | 0.7333 | 0.2667 | 0.4667 |
| B | 0.0667 | 0.0167 | 0.0333 | B | 0.1500 | 0.4000 | 0.2333 | ||
| C | 0.400 | 0.3000 | 0.4833 | C | 0.1167 | 0.3333 | 0.3000 | ||
| D | 0.0333 | 0.2167 | 0.0667 | ||||||
| TUZXPM49 | A | 0.0500 | 0.0333 | 0.0167 | PM1382 | A | 0.3000 | 0.3000 | 0.4667 |
| B | 0.5167 | 0.5667 | 0.4667 | B | 0.3000 | 0.0833 | 0.0500 | ||
| C | 0.4333 | 0.4000 | 0.5167 | C | 0.4000 | 0.6167 | 0.4833 | ||
| PM1713 | A | 0.7333 | 0.6500 | 0.7333 | DTLPM104 | A | 0.2333 | 0.1000 | 0.0500 |
| B | 0.2667 | 0.3500 | 0.2667 | B | 0.6833 | 0.7833 | 0.6333 | ||
| C | 0.0833 | 0.1167 | 0.3167 | ||||||
| BC52 | A | 0.3167 | 0.5833 | 0.0667 | PM1381 | A | 0.6500 | 0.4667 | 0.3833 |
| B | 0.6833 | .4167 | 0.9333 | B | 0.3500 | 0.5333 | 0.6167 | ||
| PM528 | A | 0.3167 | 0.1500 | 0.3167 | PM138 | A | 0.0500 | — | 0.0667 |
| B | 0.5333 | 0.6333 | 0.5167 | B | 0.6833 | 0.9333 | 0.8333 | ||
| C | 0.1500 | 0.2167 | 0.1667 | C | 0.2667 | 0.0667 | 0.1000 | ||
| PM2345 | A | 0.0833 | 0.0667 | 0.0167 | PM3852 | A | 0.1667 | 0.1833 | 0.1333 |
| B | 0.8667 | 0.9333 | 0.9333 | B | 0.5833 | 0.6500 | 0.6333 | ||
| C | 0.0500 | — | 0.0500 | C | 0.2500 | 0.1667 | 0.2333 | ||
| PM205 | A | 0.6167 | 0.6167 | 0.6667 | PM4793 | A | 0.5167 | 0.4333 | 0.7500 |
| B | 0.3833 | 0.3833 | 0.3333 | B | 0.4833 | 0.5667 | 0.2500 | ||
Tab. 3
Genetic variation of three populations (Qionghai, Sanya, and Zhanjiang) of P. monodon"
| 位点 | 等位 基因数 | 有效等位 基因数 | 观察杂合度 | 期望杂合度 | HW平衡偏离指数 | HW 平衡检测 | 多态信息含量 |
|---|---|---|---|---|---|---|---|
| DTLPM120 | 4/4/4 | 2.4064/2.8169/2.4226 | 1.000/0.9333/0.9333 | 0.5844/0.6450/0.5872 | 0.7111/0.4470/0.5894 | **/**/* | 0.5804/0.6352/0.5842 |
| TUZXPM49 | 3/3/3 | 2.1871/2.0737/2.0619 | 0.4667/0.6667/0.4000 | 0.5428/0.5178/0.5150 | -0.1402/0.2876/-0.2233 | ns/ns/ns | 0.4212/0.4162/0.3985 |
| PM1713 | 2/2/2 | 1.6423/1.8349/1.6423 | 0.2667/0.5667/0.4667 | 0.3911/0.4550/0.3911 | -0.3181/0.2455/0.1933 | ns/ns/ns | 0.3146/0.3515/0.3146 |
| BC52 | 2/2/2 | 1.7630/1.9459/1.1421 | 0.5667/0.3667/0.1333 | 0.4328/0.4861/0.1244 | 0.3094/-0.2456/-0.0715 | ns/ns/ns | 0.3391/0.3668/0.1168 |
| DTLPM402 | 3/3/3 | 1.7425/2.9221/2.7607 | 0.3333/0.3000/0.4000 | 0.4261/0.6578/0.6378 | -0.2178/-0.5439/-0.3728 | ns/**/* | 0.4013/0.5995/0.6043 |
| DTLPM303 | 3/3/3 | 2.9412/2.0955/2.2032 | 0.5000/0.6333/0.4000 | 0.6600/0.5228/0.5461 | -0.2424/0.2114/-0.2675 | ns/ns/ns | 0.51/0.5162/0.5439 |
| DTLPM104 | 3/3/3 | 1.8927/1.5693/1.9846 | 0.4667/0.4333/0.4667 | 0.4717/0.3628/0.4961 | -0.011/0.1943/-0.0593 | ns/ns/ns | 0.4146/0.3338/0.4137 |
| DTLPM218 | 2/2/2 | 1.8349/1.9912/1.8967 | 0.4333/0.4667/0.2333 | 0.4550/0.4978/0.4728 | -0.0478/-0.0625/-0.5066 | ns/ns/** | 0.3515/0.3739/0.3857 |
| PM528 | 3/3/3 | 2.4557/2.1251/2.5316 | 0.4000/0.3000/0.4333 | 0.5928/0.5294/0.6050 | -0.3252/-0.4333/-0.2838 | */**/ns | 0.5231/0.4738/0.5365 |
| PM205 | 2/2/2 | 1.8967/1.8967/1.8000 | 0.3667/0.6333/0.6000 | 0.4728/0.4728/0.4444 | -0.2244/0.3395/0.3501 | ns/ns/ns | 0.3610/0.3610/0.3457 |
| PM138 | 3/2/3 | 1.8499/1.1421/1.4107 | 0.4333/0.0000/0.1333 | 0.4594/0.1244/0.2911 | -0.0568/-1.000/-0.5421 | ns/**/** | 0.3902/0.1168/0.2711 |
| PM3852 | 3/3/3 | 2.3226/2.0666/2.1127 | 0.3667/0.2333/0.2333 | 0.5694/0.5161/0.5267 | -0.3559/-0.5480/-0.5571 | */**/** | 0.5025/0.4642/0.4688 |
| PM2345 | 3/2/3 | 1.3148/1.1421/1.1443 | 0.1667/0.0667/0.0667 | 0.2394/0.1244/0.1261 | -0.3037/-0.4638/-0.4711 | **/*/** | 0.2252/0.1168/0.1213 |
| PM4793 | 2/2/2 | 1.9978/1.9651/1.6000 | 0.1000/0.5333/0.2333 | 0.4994/0.4911/0.3750 | -0.7998/0.0860/-0.3787 | **/ns/* | 0.3747/0.3705/0.3047 |
| 平均 | 2.7143/ 2.5714/ 2.7143 | 2.0185/1.9715/1.9081 | 0.4214/0.4357/0.3667 | 0.4857/0.4575/0.4385 | -0.2926/-0.2809/-0.2817 | ns/ns/ns | 0.4078/0.3926/0.3864 |
Tab. 5
F-value and gene flow of 14 microsatellite loci in the three populations of P. monodon"
| 微卫星位点 | 近交系数 | 遗传分化系数 | 基因流 | 微卫星位点 | 近交系数 | 遗传分化系数 | 基因流 |
|---|---|---|---|---|---|---|---|
| DTLPM120 | -0.5780 | 0.0219 | 11.1477 | DTLPM402 | 0.4197 | 0.0887 | 2.5679 |
| TUZXPM49 | 0.0070 | 0.0079 | 31.3235 | DTLPM218 | 0.2050 | 0.0496 | 4.7873 |
| PM1713 | -0.0507 | 0.0074 | 33.4050 | DTLPM303 | 0.1131 | 0.0438 | 5.4533 |
| BC52 | -0.0224 | 0.2038 | 0.9768 | DTLPM104 | -0.0271 | 0.0442 | 5.4104 |
| PM528 | 0.3438 | 0.0165 | 14.9471 | PM3852 | 0.4831 | 0.0047 | 53.0854 |
| PM205 | -0.1511 | 0.0024 | 104.2500 | PM2345 | 0.3878 | 0.0142 | 17.4079 |
| PM138 | 0.3524 | 0.0612 | 3.8352 | PM4793 | 0.3653 | 0.0732 | 3.1675 |
| 平均 | 0.1143 | 0.0456 | 5.2321 |
| [1] | 杜晓东, 秦红贵, 黄荣莲, 等, 2004. 斑节对虾两个种群生化遗传变异的研究[J]. 海洋科学, 28(6): 32-36. |
| DU XIAODONG, QIN HONGGUI, HUANG RONGLIAN, et al, 2004. Studies on biochemical genetic variation of two populations of Penaeus monodon Fabricius[J]. Marine Science, 28(6): 32-36 (in Chinese). | |
| [2] | 姜永杰, 周发林, 黄键华, 等, 2006. 深圳海域斑节对虾野生种群线粒体控制区序列的多态性分析[J]. 南方水产, 2(1): 54-57. |
| JIANG YONGJIE, ZHOU FALIN, HUANG JIANHUA, et al, 2006. Analysis on genetic diversity of mtDNA control region sequences of the wild population of Peneanus monodon from Shenzhen's sea area[J]. South China Fisheries Science, 2(1): 54-57 (in Chinese). | |
| [3] | 姜因萍, 何毛贤, 黄良民, 等, 2008. 两个大珠母贝群体遗传多样性的ISSR分析[J]. 热带海洋学报, 27(3): 61-65. |
| JIANG YINPING, HE MAOXIAN, HUANG LIANGMIN, et al, 2008. ISSR analysis of genetic diversity in two populations of pearl oyster Pinctada maxima[J]. Journal of Tropical Oceanography, 27(3): 61-65 (in Chinese). | |
| [4] | 李康, 杜晓东, 叶富良, 2005. 斑节对虾两个野生种群RAPD分析[J]. 湛江海洋大学学报, 25(3): 79-82. |
| [5] | 钱迎倩, 马克平, 1994. 生物多样性研究的原理与方法[M]. 北京: 中国科学技术出版社. |
| [6] | 苏天凤, 熊小飞, 江世贵, 等, 2010. 斑节对虾7个全同胞家系间亲缘关系的微卫星分析[J]. 南方水产, 6(6): 1-7. |
| SU TIANFENG, XIONG XIAOFEI, JIANG SHIGUI, et al, 2010. Microsatellite analysis on genetic relationships among 7 full-sib families of Penaeus monodon[J]. South China Fisheries Science, 6(6): 1-7 (in Chinese). | |
| [7] | 谭树华, 王桂忠, 艾春香, 等, 2005. 斑节对虾养殖群体遗传多样性传多样性的同工酶和RAPD分析[J]. 中国水产科学, 12(6): 702-707. |
| TAN SHUHUA, WANG GUIZHONG, AI CHUNXIANG, et al, 2005. Genetic diversity of reared Penaeus monodon stock revealed by isozyme and RAPD[J]. Journal of Fishery Sciences of China, 12(6): 702-707 (in Chinese). | |
| [8] | 谭树华, 王桂忠, 李少菁, 2011. 斑节对虾两个野生亲虾种群遗传多样性[J]. 生态学杂志, 30(9): 2002-2006. |
| TAN SHUHUA, WANG GUIZHONG, LI SHAOJING, 2011. Genetic diversity of two wild Penaeus monodon broodstock populations[J]. Chinese Journal of Ecology, 30(9): 2002-2006 (in Chinese). | |
| [9] | 熊小飞, 夏军红, 苏天凤, 等, 2008. 海南、深圳和北海斑节对虾群体遗传多样性研究[J]. 水利渔业, 28(3): 30-31, 38. |
| [10] | 周发林, 江世贵, 姜永杰, 等, 2006. 海南三亚斑节对虾野生种群mtDNA 16S rRNA基因和控制区序列的多态性[J]. 南方水产, 2(6): 13-18. |
| ZHOU FALIN, JIANG SHIGUI, JIANG YONGJIE, et al, 2006. Polymorphism of mtDNA 16S rRNA gene and control region sequence in Penaeus monodon of Sanya, Hainan[J]. South China Fisheries Science, 2(6): 13-18 (in Chinese). | |
| [11] | 周发林, 江世贵, 姜永杰, 等, 2009. 中国南海野生斑节对虾5个地理群体线粒体16S rRNA基因序列比较分析[J]. 水产学报, 33(2): 208-213. |
| ZHOU FALIN, JIANG SHIGUI, JIANG YONGJIE, et al, 2009. Assessing genetic diversity of the five wild tiger prawn (Penaeus monodon) populations in the coastal waters of South China Sea, based on mitochondrial DNA 16S rRNA sequences[J]. Journal of Fisheries of China, 33(2): 208-213 (in Chinese). | |
| [12] | BENZIE J A H, 2000. Population genetic structure in penaeid prawns[J]. Aquaculture Research, 31(1): 95-119. |
| [13] | BOTSTEIN D, WHITE R L, SKOLNICK M, et al, 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms[J]. American Journal of Human Genetics, 32(3): 314-331. |
| [14] | EXCOFFIER L, SMOUSE P E, QUATTRO J M, 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data[J]. Genetics, 131(2): 479-491. |
| [15] | HAMRICK J L, GODT M J W, SHERMAN-BROYLES S L, 1995. Gene flow among plant populations: evidence from genetic markers[C]// HOCH P C, STEPHENSON A G. Experimental and molecular approaches to plant biosystematics. St. Louis: Missouri Botanical Garden Press: 215-232. |
| [16] | LUCIEN-BRUN H, 1997. Evolution of world shrimp production: Fisheries and aquaculture[J]. World Aquaculture, 28: 21-33. |
| [17] | PAN Y W, CHOU H H, YOU E M, et al, 2004. Isolation and characterization of 23 polymorphic microsatellite markers for diversity and stock analysis in tiger shrimp (Penaeus monodon)[J]. Molecular Ecology Notes, 4(3): 345-347. |
| [18] | SAMBROOK J, RUSSELL D W, 2008. Molecular Cloning: a laboratory manual[M]. 3rd ed. Cold Spring Harbour, New York: Cold Spring Harbor Laboratory Press. |
| [19] | Slatkin M, 1985. Rare alleles as indicators of gene flow[J]. Evolution, 39(1): 53-65. |
| [20] | SOKAL R R, JACQUEZ G M, WOOTEN M C, 1989. Spatial autocorrelation analysis of migration and selection[J]. Genetics, 121(4): 845-855. |
| [21] | SUGAMA K, HARYANTI, BENZIE J A H, et al, 2002. Genetic variation and population structure of the giant tiger prawn, Penaeus monodon, in Indonesia[J]. Aquaculture, 205(1-2): 37-48. |
| [22] | SUPUNGUL P, SOOTANAN P, KLINBUNGA S, et al, 2000. Microsatellite polymorphism and the population structure of the black tiger shrimp (Penaeus monodon) in Thailand[J]. Marine Biotechnology, 2(4): 339-347. |
| [23] | WUTHISUTHIMETHAVEE S, LUMUBOL P, VANAVICHIT A, et al, 2003. Development of microsatellite markers in black tiger shrimp (Penaeus monodon Fabricius)[J]. Aquaculture, 224(1-4): 39-50. |
| [24] | XU Z K, PRIMAVERA JH, DE LA PENA L D, et al, 2000. Genetic diversity of wild and cultured black tiger shrimp (Penaeus monodon) in the Philippines using microsatellites[J]. Aquaculture, 199(1-2): 13-40. |
| [1] | FU Chengchong, LI Fuyu, CHEN Dandan, HOU Jing, WANG Jun, LI Yuanchao, WANG Daoru, WANG Yan. The genetic structure and connectivity of Porites lutea metapopulation of the fringing reefs around the Hainan Island [J]. Journal of Tropical Oceanography, 2023, 42(2): 64-77. |
| [2] | WU Weizhi, ZHAO Zhixia, YANG Sheng, LIANG Licheng, Chen Qiuxia, LU Xiang, LIU Xing, ZHANG Xiaowei. The mangrove forest distribution and analysis of afforestation effect in Zhejiang Province [J]. Journal of Tropical Oceanography, 2022, 41(6): 67-74. |
| [3] | YANG Liuqinqing, CHU Moxian, LIU Bilin, KONG Xianghong. Pattern recognition of Sthenoteuthis oualaniensis based on BPNN about momentum and self-adaption [J]. Journal of Tropical Oceanography, 2021, 40(6): 102-110. |
| [4] | ZOU Congcong, WANG Lijuan, WU Zhihao, YOU Feng. Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences* [J]. Journal of Tropical Oceanography, 2021, 40(5): 25-35. |
| [5] | REN Huimin, ZHANG Heng, XU Shasha, LI Shengfa, LI Jiansheng, LI Zhihong, HE Lijun. Population genetic structure and historical dynamics of Trachurus japonicus in the China seas based on mitochondrial control region [J]. Journal of Tropical Oceanography, 2021, 40(5): 36-44. |
| [6] | Min LI, Xiaolan KONG, Youwei XU, Zuozhi CHEN. Genetic polymorphism of the Brushtooth lizardfish Saurida undosquamis based on mitochondrial D-loop sequences [J]. Journal of Tropical Oceanography, 2020, 39(4): 42-49. |
| [7] | Lingli WANG, Simin HU, Minglan GUO, Tao LI, Youjun WANG, Hui HUANG, Sheng LIU. In situ feeding differences between adults and juveniles of chaetognath (Flaccisagitta enflata) in Sanya Bay [J]. Journal of Tropical Oceanography, 2020, 39(3): 57-65. |
| [8] | Hui WANG,Hengxiang LI,Lu LI,Yan YAN. The population distribution of Hyale grandicornis in macroalgae canopies of Daya Bay [J]. Journal of Tropical Oceanography, 2019, 38(4): 52-58. |
| [9] | Yingying JI,Lei XU,Hong LI,Lianggen WANG,Feiyan DU. Genetic structure of Oithona setigera from South China Sea based on 28S rDNA gene [J]. Journal of Tropical Oceanography, 2019, 38(3): 89-97. |
| [10] | Xiaolin HUANG, Wenjun LI, Heizhao LIN, Yukai YANG, Tao LI, Wei YU, Zhong HUANG. Genetic variations among Siganus oramin populations in coastal waters of southeast China based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2018, 37(4): 45-51. |
| [11] | ZHU Kai, WANG Xuehui, ZHANG Peng, DU Feiyan, QIU Yongsong. A study on morphological variations and discrimination of medium and dwarf forms of purple flying squid Sthenoteuthis oualaniensis in the southern South China Sea [J]. Journal of Tropical Oceanography, 2016, 35(6): 82-88. |
| [12] | ZHANG Kui, CHEN Zuozhi, WANG Yuezhong, SUN Dianrong, QIU Yongsong. Population structure of Priacanthus macracanthus in the Beibu Gulf, and parameters for its growth, mortality and maturity [J]. Journal of Tropical Oceanography, 2016, 35(5): 20-28. |
| [13] | WENG Zhaohong, XIE Yangjie, XIAO Zhiqun, WANG Yilei. Analysis of genetic diversity in several wild and hatchery populations of Crassostrea angulata from south Fujian and south Guangdong [J]. Journal of Tropical Oceanography, 2016, 35(3): 94-98. |
| [14] | SHI Xiao-feng1, 2, SU Yong-quan1, WANG Wen-cheng1, WANG Jun1. Population genetic structure of three stocks of Acanthopagrus schlegelii based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2015, 34(1): 56-63. |
| [15] | LIU Li, ZHAO Jie, GUO Yu-song, LIU Chu-wu. Study on the AFLP primer selection and genetic diversities of 4 Snappers [J]. Journal of Tropical Oceanography, 2012, 31(4): 112-116. |
|
||