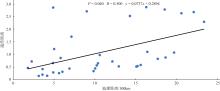
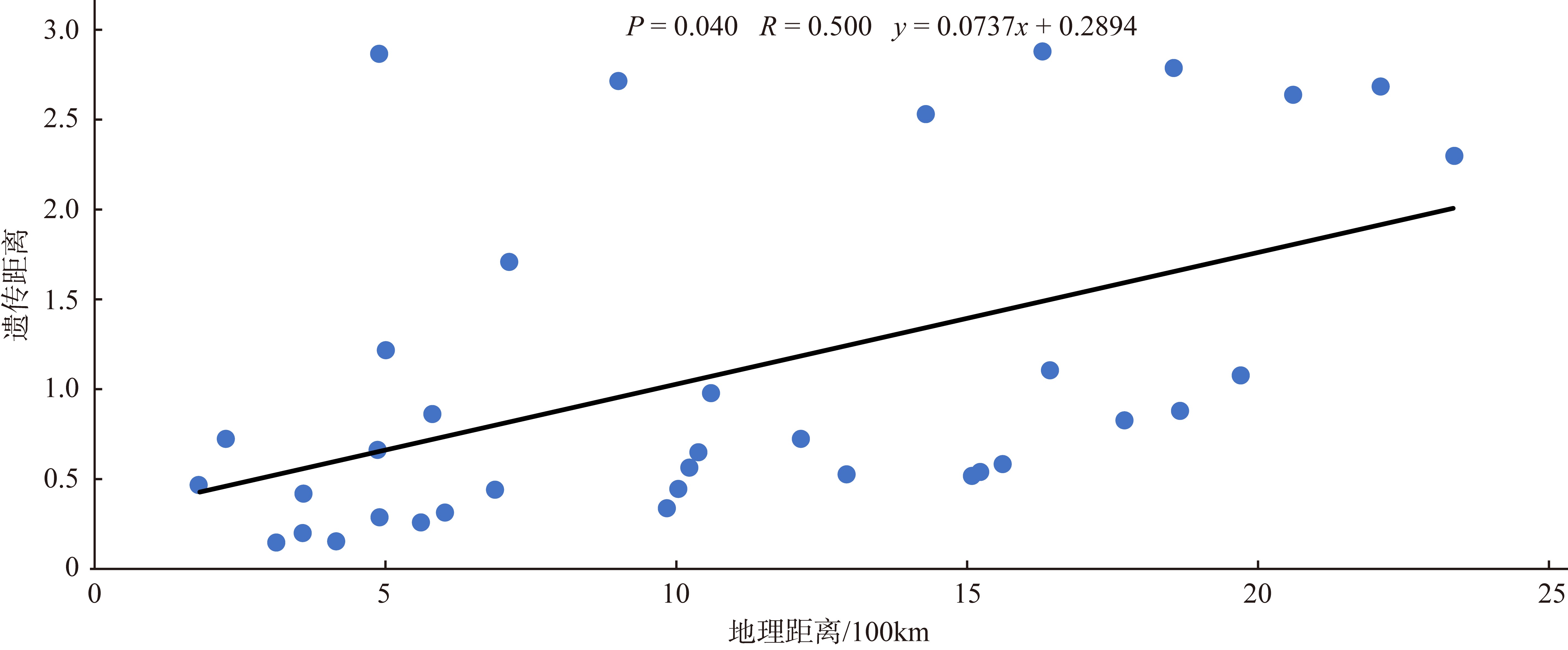
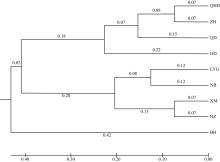
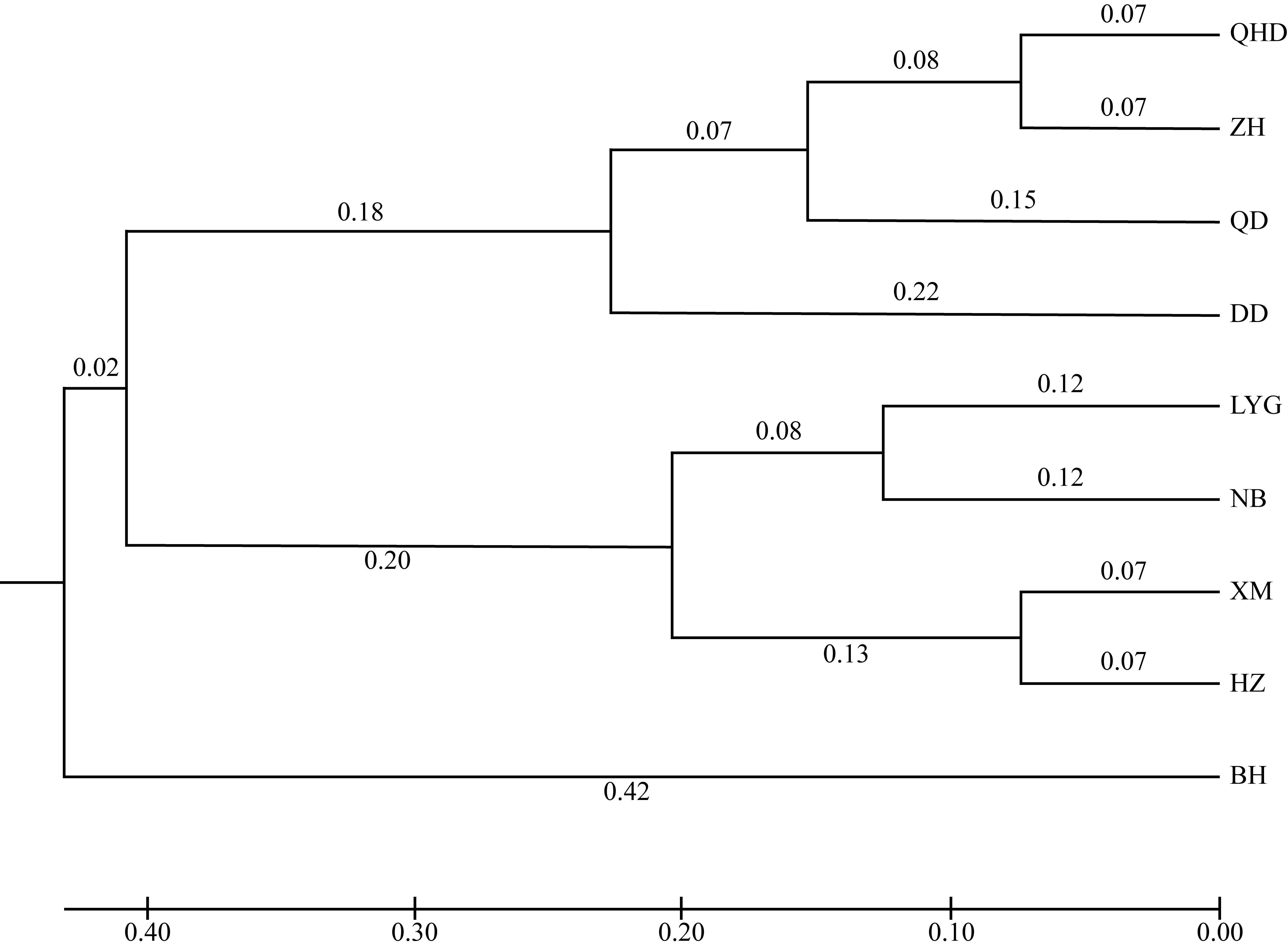
Journal of Tropical Oceanography ›› 2025, Vol. 44 ›› Issue (2): 124-136.doi: 10.11978/2024117CSTR: 32234.14.2024117
• Marine Biology • Previous Articles Next Articles
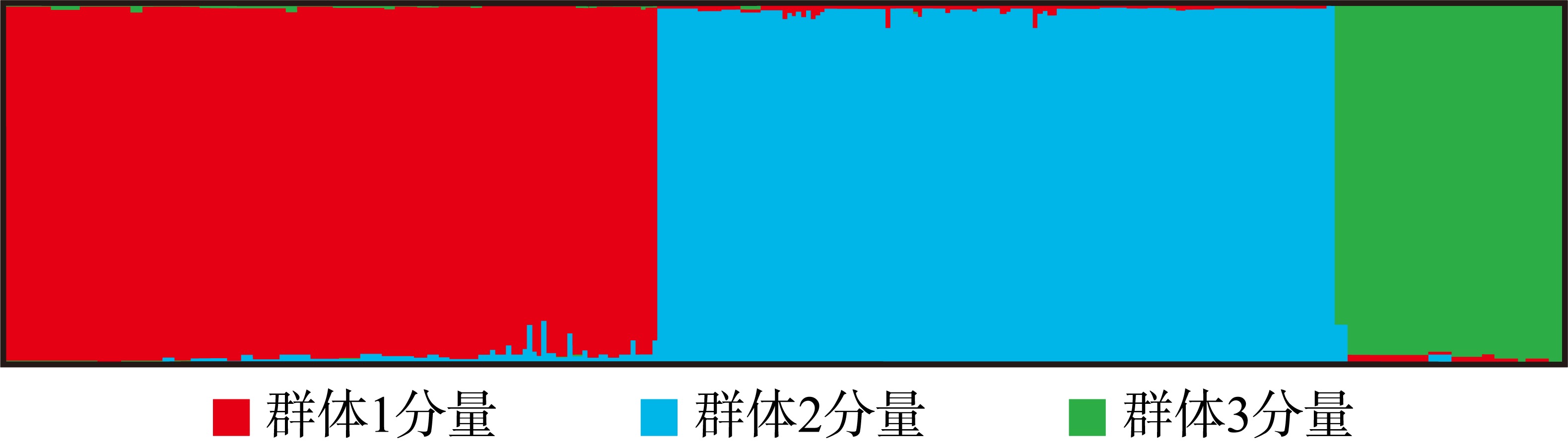
Analysis of genetic diversity in 9 populations of Sinonovacula constricta using microsatellite markers
WU Guiqing1( ), LI Ruihua1, XIAO Yihao1, CHEN Yanlin1, LUO Xuan2, LIU Xiangquan3, ZHU Jiajie1, WU Xueping1(
), LI Ruihua1, XIAO Yihao1, CHEN Yanlin1, LUO Xuan2, LIU Xiangquan3, ZHU Jiajie1, WU Xueping1( )
)
- 1. School of Marine Science and Biotechnology, Guangxi Minzu University, Nanning 530007, China
2. Guangxi Fisheries Technical Extension Station, Nanning 530016, China
3. Shandong Provincial Key Laboratory of Restoration for Marine Ecology (Shandong Marine Resource and Environment Research Institute), Yantai 264006, China
-
Received:2024-06-01Revised:2024-08-19Online:2025-03-10Published:2025-04-11 -
Contact:WU Xueping -
Supported by:High Level Introduction of Talent Research Start-up Projects of Guangxi Minzu University(2017KJQD007); Guangxi Natural Science Foundation Grant(2020GXNSFBA159010); Innovation and Entrepreneurship Training Program for College Students of Guangxi in 2022(S202210608162); Innovation and Entrepreneurship Training Program for College Students of Guangxi in 2022(S202210608163); Innovation and Entrepreneurship Training Program for College Students of Guangxi in 2023(S202310608040)
CLC Number:
- P735.542
Cite this article
WU Guiqing, LI Ruihua, XIAO Yihao, CHEN Yanlin, LUO Xuan, LIU Xiangquan, ZHU Jiajie, WU Xueping. Analysis of genetic diversity in 9 populations of Sinonovacula constricta using microsatellite markers[J].Journal of Tropical Oceanography, 2025, 44(2): 124-136.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Sampling information of 9 populations of S. constricta"
| 采样地点 | 经度 | 纬度 | 样本量/个 | 采样时间 |
|---|---|---|---|---|
| DD | 124°16′54′′E | 39°56′15′′N | 30 | 2022年11月22日 |
| QHD | 119°26′11′′E | 39°47′47′′N | 30 | 2022年11月05日 |
| ZH | 122°38′59′′E | 39°31′36′′N | 30 | 2022年10月23日 |
| QD | 120°67′16′′E | 36°16′06′′N | 30 | 2022年10月15日 |
| LYG | 119°17′08′′E | 34°46′50′′N | 30 | 2022年9月27日 |
| NB | 121°43′55′′E | 29°32′01′′N | 30 | 2022年9月18日 |
| XM | 118°11′50′′E | 24°29′18′′N | 30 | 2022年8月25日 |
| HZ | 114°38′26′′E | 22°45′40′′N | 30 | 2022年8月12日 |
| BH | 109°06′33′′E | 21°25′25′′N | 30 | 2022年7月10日 |
Tab. 2
Information of 10 pairs of microsatellite locus of S. constricta"
| 位点 | 序列号 | 引物序列(5′-3′) | 退火温度/℃ | 片段大小/bp |
|---|---|---|---|---|
| SC03 | PP409554 | F: CTTATCTTTATCCTCCTCGCCAT R: TGATGATAATGTTGAGGCTGTTG | 54 | 120~160 |
| SC04 | PP409555 | F: AGATAAAGCCAGGAAAACACAGA R: GTTTACAATGAAAAGACAGATGGC | 52 | 110~150 |
| SC05 | PP409556 | F: TCTTGGTTAGAGTGGGTTGTTGT R: TGCTTTCTAATGTGCCTTTCATT | 52 | 130~170 |
| SC06 | PP409557 | F: TTTCATGTTGTTGGTTGTTTGTT R: AATGTTGCCAAATCAAAACATTC | 54 | 110~150 |
| SC07 | PP409558 | F: CATCATCATTAACCGTCCTCCTA R:AGAAATTGGCATTGATAGCAAAA | 52 | 120~160 |
| SC08 | PP409559 | F: CCATGCATGAGAGTTAAGTAACAAA R: CCAAGAATGCACAATAAAATTGA | 52 | 120~160 |
| SC11 | PP409560 | F: GACAATTATGATTGCGATTGGTT R: TTGATACCCTTCATTCCAGCTTA | 53 | 120~150 |
| SC13 | PP409560 | F: ACAACGCCAGCAGTAACCTATAA R: CGGTTGTGGAAGCAGTAGTAGTAG | 54 | 100~140 |
| SC17 | PP409562 | F: CACCTCTGTCTGAAAAACAAAAA R: AGGGACACCCAGTACACATCTAT | 50 | 130~170 |
| SC19 | PP409563 | F: CAAAACCGGTTGGTAATGAATAA R: TTTTGTTGTTGTTTTGTTGTTGC | 55 | 120~160 |
Tab. 3
Average genetic diversity index of 9 populations of S. constricta"
| 群体 | 参数 | SC03 | SC04 | SC05 | SC06 | SC07 | SC08 | SC11 | SC13 | SC17 | SC19 | 均值 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DD | Na | 3.0000 | 3.0000 | 4.0000 | 5.0000 | 6.0000 | 5.0000 | 4.0000 | 4.0000 | 4.0000 | 4.0000 | 4.2000 |
| Ne | 1.7527 | 1.9674 | 2.3810 | 4.3239 | 2.9268 | 3.4118 | 3.2200 | 1.8127 | 1.4207 | 2.2161 | 2.5433 | |
| Ho | 0.6000 | 0.0357 | 0.1000 | 0.4138 | 0.3333 | 0.6207 | 0.7667 | 0.2667 | 0.2333 | 0.0690 | 0.3439 | |
| He | 0.4367 | 0.5006 | 0.5898 | 0.7822 | 0.6695 | 0.7193 | 0.7011 | 0.4559 | 0.3011 | 0.5584 | 0.5715 | |
| PIC | 0.4510 | 0.4130 | 0.7330 | 0.7730 | 0.6070 | 0.9080 | 0.6650 | 0.4390 | 0.2870 | 0.4580 | 0.5734 | |
| P-value | 1.0000 | 0.0000* | 0.0000* | 0.0000* | 0.0033 | 0.0042 | 0.8245 | 0.0002* | 0.0561 | 0.0000* | 0.1888 | |
| QHD | Na | 5.0000 | 3.0000 | 5.0000 | 5.0000 | 4.0000 | 5.0000 | 3.0000 | 5.0000 | 4.0000 | 2.0000 | 4.1000 |
| Ne | 2.2416 | 1.4551 | 3.5644 | 4.4888 | 2.9851 | 3.3835 | 2.0619 | 1.6949 | 1.2721 | 1.9627 | 2.5110 | |
| Ho | 0.7333 | 0.0333 | 0.5333 | 0.3667 | 0.4667 | 0.6000 | 0.9667 | 0.4000 | 0.2333 | 0.0345 | 0.4368 | |
| He | 0.5633 | 0.3181 | 0.7316 | 0.7904 | 0.6763 | 0.7164 | 0.5237 | 0.4169 | 0.2175 | 0.4991 | 0.5453 | |
| PIC | 0.5690 | 0.2860 | 0.7040 | 0.7550 | 0.6230 | 0.8260 | 0.3980 | 0.4050 | 0.2050 | 0.3700 | 0.5141 | |
| P-value | 0.9964 | 0.0000* | 0.0000* | 0.0641 | 0.0025 | 1.0000 | 0.0459 | 1.0000 | 0.0000* | 0.0205 | 0.3129 | |
| ZH | Na | 4.0000 | 3.0000 | 6.0000 | 5.0000 | 4.0000 | 6.0000 | 3.0000 | 5.0000 | 3.0000 | 3.0000 | 4.2000 |
| Ne | 2.5388 | 1.1450 | 2.9561 | 4.1570 | 2.5678 | 4.5113 | 2.3866 | 1.9759 | 1.2685 | 1.4019 | 2.4909 | |
| Ho | 0.2667 | 0.0000 | 0.4138 | 0.3667 | 0.2000 | 0.6000 | 0.6071 | 0.4000 | 0.2333 | 0.0000 | 0.3088 | |
| He | 0.6164 | 0.1288 | 0.6733 | 0.7723 | 0.6209 | 0.7915 | 0.5916 | 0.5023 | 0.2153 | 0.2915 | 0.5204 | |
| PIC | 0.5360 | 0.1230 | 0.6750 | 0.7330 | 0.5300 | 0.8940 | 0.5080 | 0.4630 | 0.1990 | 0.2600 | 0.4921 | |
| P-value | 0.0205 | 0.0003* | 0.0022 | 0.0000* | 0.0001* | 0.0000* | 0.0386 | 0.2979 | 1.0000 | 0.0000* | 0.1359 | |
| QD | Na | 5.0000 | 2.0000 | 4.0000 | 5.0000 | 3.0000 | 4.0000 | 3.0000 | 5.0000 | 2.0000 | 4.0000 | 3.7000 |
| Ne | 1.6559 | 1.0689 | 3.4682 | 4.5421 | 1.3072 | 3.4221 | 2.0202 | 1.3206 | 1.0339 | 1.6408 | 2.1480 | |
| Ho | 0.4000 | 0.0000 | 0.5333 | 0.2963 | 0.2000 | 0.7000 | 0.8667 | 0.2000 | 0.0333 | 0.1333 | 0.3363 | |
| He | 0.4028 | 0.0655 | 0.7237 | 0.7945 | 0.239 | 0.7198 | 0.5136 | 0.2469 | 0.0333 | 0.3972 | 0.4136 | |
| PIC | 0.3770 | 0.0620 | 0.0730 | 0.7560 | 0.2220 | 0.8590 | 0.3930 | 0.2340 | 0.0320 | 0.3650 | 0.3373 | |
| P-value | 0.0316 | 0.0169 | 0.0036 | 0.0000* | 0.2409 | 0.0000* | 1.0000 | 0.0146 | 0.0000* | 0.0002* | 0.1308 | |
| LYG | Na | 1.0000 | 1.0000 | 5.0000 | 2.0000 | 2.0000 | 6.0000 | 4.0000 | 5.0000 | 3.0000 | 3.0000 | 3.2000 |
| Ne | 1.0000 | 1.0000 | 1.9956 | 1.2596 | 1.9978 | 4.2254 | 3.1634 | 1.7787 | 1.2237 | 1.1443 | 1.8789 | |
| Ho | 0.0000 | 0.0000 | 0.3667 | 0.2333 | 0.9667 | 0.7333 | 0.6333 | 0.3667 | 0.1333 | 0.0667 | 0.3500 | |
| He | 0.0000 | 0.0000 | 0.5073 | 0.2096 | 0.5079 | 0.7763 | 0.6955 | 0.4452 | 0.1859 | 0.1282 | 0.3456 | |
| PIC | 0.0000 | 0.0320 | 0.6650 | 0.1850 | 0.3750 | 0.8710 | 0.6190 | 0.4160 | 0.1800 | 0.1240 | 0.3467 | |
| P-value | 0.0000* | 1.0000 | 0.0000* | 1.0000 | 1.0000 | 0.1941 | 0.2597 | 0.0006* | 0.0669 | 0.0169 | 0.3538 | |
| NB | Na | 5.0000 | 2.0000 | 5.0000 | 5.0000 | 2.0000 | 5.0000 | 3.0000 | 5.0000 | 2.0000 | 4.0000 | 3.8000 |
| Ne | 3.1746 | 1.0689 | 3.3835 | 3.1142 | 1.4274 | 4.0909 | 2.2727 | 2.2814 | 1.0339 | 1.4718 | 2.3319 | |
| Ho | 0.4667 | 0.0000 | 0.4667 | 0.4333 | 0.3667 | 0.5333 | 0.9333 | 0.4000 | 0.0333 | 0.1000 | 0.3733 | |
| He | 0.6966 | 0.0655 | 0.7164 | 0.6904 | 0.3045 | 0.7684 | 0.5695 | 0.5712 | 0.0333 | 0.3260 | 0.4742 | |
| PIC | 0.6560 | 0.0620 | 0.8160 | 0.6430 | 0.2550 | 0.9030 | 0.4610 | 0.5500 | 0.0640 | 0.3050 | 0.4715 | |
| P-value | 0.1371 | 0.0169 | 0.0000* | 0.0058 | 1.0000 | 0.0217 | 0.9827 | 0.0000* | 1.0000 | 0.0000* | 0.3164 | |
| XM | Na | 2.0000 | 1.0000 | 4.0000 | 5.0000 | 5.0000 | 5.0000 | 3.0000 | 4.0000 | 4.0000 | 5.0000 | 3.8000 |
| Ne | 1.6000 | 1.0000 | 2.2876 | 3.5573 | 1.9063 | 3.8793 | 2.2032 | 2.1102 | 1.4634 | 1.8653 | 2.1873 | |
| Ho | 0.4333 | 0.0000 | 0.3462 | 0.3667 | 0.4333 | 0.8000 | 0.9333 | 0.4333 | 0.2000 | 0.1000 | 0.4046 | |
| He | 0.3814 | 0.0000 | 0.5739 | 0.7311 | 0.4836 | 0.7548 | 0.5554 | 0.5350 | 0.3220 | 0.4718 | 0.4809 | |
| PIC | 0.6720 | 0.4410 | 0.8840 | 0.4420 | 0.4410 | 0.8840 | 0.4420 | 0.5260 | 0.2950 | 0.4440 | 0.5471 | |
| P-value | 0.0000* | 0.0044 | 0.6226 | 0.8616 | 0.0044 | 0.6226 | 0.8616 | 0.0068 | 0.0048 | 0.0000* | 0.2989 | |
| HZ | Na | 5.0000 | 3.0000 | 5.0000 | 5.0000 | 2.0000 | 5.0000 | 4.0000 | 4.0000 | 2.0000 | 4.0000 | 3.9000 |
| Ne | 2.2939 | 1.2649 | 2.4931 | 3.4417 | 1.5139 | 3.8710 | 2.8257 | 1.3657 | 1.0339 | 1.5139 | 2.1618 | |
| Ho | 0.5333 | 0.0333 | 0.2333 | 0.5667 | 0.4333 | 0.6333 | 0.7667 | 0.3000 | 0.0333 | 0.1333 | 0.3667 | |
| He | 0.5734 | 0.2130 | 0.6090 | 0.7215 | 0.3452 | 0.7542 | 0.6571 | 0.2723 | 0.0333 | 0.3452 | 0.4524 | |
| PIC | 0.5880 | 0.2130 | 0.7830 | 0.7260 | 0.3520 | 0.9010 | 0.6570 | 0.2450 | 0.0330 | 0.3460 | 0.4844 | |
| P-value | 0.0937 | 0.0001* | 0.0000* | 0.0683 | 1.0000 | 0.0000* | 0.6531 | 1.0000 | 0.0000* | 0.0263 | 0.2841 | |
| BH | Na | 5.0000 | 3.0000 | 5.0000 | 3.0000 | 5.0000 | 5.0000 | 5.0000 | 5.0000 | 3.0000 | 4.0000 | 4.3000 |
| Ne | 2.2032 | 1.2270 | 2.4658 | 1.3943 | 4.5226 | 1.8927 | 2.8939 | 2.8571 | 1.9231 | 1.7561 | 2.3136 | |
| Ho | 0.6333 | 0.2000 | 0.4333 | 0.3333 | 0.7667 | 0.5333 | 0.6667 | 0.7667 | 0.6667 | 0.5333 | 0.5533 | |
| He | 0.5554 | 0.1881 | 0.6045 | 0.2876 | 0.7921 | 0.4797 | 0.6655 | 0.6610 | 0.4881 | 0.4379 | 0.5160 | |
| PIC | 0.5030 | 0.1770 | 0.7760 | 0.2510 | 0.8590 | 0.6010 | 0.7320 | 0.7160 | 0.6220 | 0.7520 | 0.5989 | |
| P-value | 0.9440 | 1.0000 | 0.0181 | 1.0000 | 0.4223 | 0.7689 | 0.9183 | 1.0000 | 0.9940 | 0.9761 | 0.8042 |
Tab. 4
Genetic differentiation index (below diagonal) and gene flow (above diagonal) of 9 populations of S. constricta"
| 群体 | DD | QHD | ZH | QD | LYG | NB | XM | HZ | BH |
|---|---|---|---|---|---|---|---|---|---|
| DD | — | 2.7297 | 1.6497 | 1.0460 | 0.5298 | 0.8741 | 0.8756 | 0.8188 | 0.8586 |
| QHD | 0.0839 | — | 4.3204 | 1.5281 | 0.6187 | 1.1282 | 1.4507 | 0.9450 | 0.7350 |
| ZH | 0.1316 | 0.0547 | — | 2.6981 | 0.7194 | 1.5093 | 1.2707 | 0.8271 | 0.8508 |
| QD | 0.1929 | 0.1406 | 0.0848 | — | 0.7186 | 1.2951 | 0.9248 | 1.0665 | 0.5406 |
| LYG | 0.3206 | 0.2878 | 0.2579 | 0.2581 | — | 1.7892 | 1.0146 | 1.0294 | 0.4620 |
| NB | 0.2224 | 0.1814 | 0.1421 | 0.1618 | 0.1226 | — | 2.1219 | 1.1939 | 0.6296 |
| XM | 0.2221 | 0.1470 | 0.1644 | 0.2128 | 0.1977 | 0.1054 | — | 3.4211 | 0.6309 |
| HZ | 0.2339 | 0.2092 | 0.2321 | 0.1899 | 0.1954 | 0.1142 | 0.0681 | — | 0.6009 |
| BH | 0.2255 | 0.2538 | 0.2721 | 0.3162 | 0.3511 | 0.2842 | 0.2838 | 0.2938 | — |
Tab. 6
Nei’s genetic distance (below diagonal) and genetic identity (above diagonal) of 9 populations"
| 群体 | DD | QHD | ZH | QD | LYG | NB | XM | HZ | BH |
|---|---|---|---|---|---|---|---|---|---|
| DD | — | 0.7501 | 0.6262 | 0.5149 | 0.1806 | 0.3759 | 0.3309 | 0.3403 | 0.1000 |
| QHD | 0.2875 | — | 0.8632 | 0.6573 | 0.2957 | 0.5220 | 0.5833 | 0.4370 | 0.0712 |
| ZH | 0.4681 | 0.1471 | — | 0.8187 | 0.4218 | 0.6405 | 0.5575 | 0.4148 | 0.0680 |
| QD | 0.6637 | 0.4196 | 0.2000 | — | 0.4845 | 0.6434 | 0.4846 | 0.5954 | 0.0613 |
| LYG | 1.7115 | 1.2185 | 0.8631 | 0.7247 | — | 0.7722 | 0.5684 | 0.5911 | 0.0559 |
| NB | 0.9785 | 0.6501 | 0.4455 | 0.4411 | 0.2586 | — | 0.7310 | 0.7133 | 0.0792 |
| XM | 1.1059 | 0.5391 | 0.5843 | 0.7244 | 0.5649 | 0.3134 | — | 0.8574 | 0.0659 |
| HZ | 1.0780 | 0.8278 | 0.8798 | 0.5185 | 0.5257 | 0.3378 | 0.1538 | — | 0.0566 |
| BH | 2.3022 | 2.6421 | 2.6885 | 2.7917 | 2.8838 | 2.5353 | 2.7198 | 2.8723 | — |
| [1] |
包文斌, 束婧婷, 许盛海, 等, 2007. 样本量和性比对微卫星分析中群体遗传多样性指标的影响[J]. 中国畜牧杂志, 43(1): 6-9.
|
|
|
|
| [2] |
范士琦, 冯婧昀, 苗晓敏, 等, 2023. 重庆养殖场鳜群体微卫星遗传多样性研究[J]. 水产养殖, 44(7): 18-23.
|
|
|
|
| [3] |
谷德贤, 王婷, 许玉甫, 等, 2021. 利用微卫星分子标记分析渤海湾的口虾蛄遗传多样性[J]. 水产科学, 40(5): 693-699.
|
|
|
|
| [4] |
郭香, 曾志南, 郑雅友, 等, 2018. 福建牡蛎选育群体的遗传多样性[J]. 中国水产科学, 25(5): 1131-1136.
|
|
|
|
| [5] |
黄小林, 李文俊, 林黑着, 等, 2018. 基于线粒体DNA D-loop序列的黄斑篮子鱼群体遗传多样性分析[J]. 热带海洋学报, 37(4): 45-51.
doi: 10.11978/2017109 |
|
|
|
| [6] |
黄新芯, 刘玉萍, 宁子君, 等, 2025. 基于微卫星标记的中国近海龙头鱼群体遗传结构分析[J]. 水生态学杂志, 46(1): 90-98.
|
|
|
|
| [7] |
李成华, 2004. 泥蚶分子系统分化及缢蛏遗传多样性的研究[D]. 青岛: 中国科学院研究生院(海洋研究所): 53-62 (in Chinese).
|
| [8] |
李景芬, 夏正龙, 栾生, 等, 2020. 五个罗氏沼虾群体遗传多样性的微卫星分析[J]. 水生生物学报, 44(6): 1208-1214.
|
|
|
|
| [9] |
李鸥, 赵莹莹, 郭娜, 等, 2009. 草鱼种群SSR分析中样本量及标记数量对遗传多度的影响[J]. 动物学研究, 30(2): 121-130.
|
|
|
|
| [10] |
李妍, 姚健涛, 张恩烁, 等, 2024. 长牡蛎(Crassostrea gigas)野生与选育群体的微卫星遗传多样性分析[J]. 海洋与湖沼, 55(2): 462-470.
|
|
|
|
| [11] |
李镒民, 丁琳琳, 谭杰, 等, 2019. 马氏珠母贝F8代黑壳色选育系群体与普通养殖群体遗传多样性分析[J]. 基因组学与应用生物学, 38(11): 4961-4967.
|
|
|
|
| [12] |
梁园, 付敬强, 沈铭辉, 等, 2022. 方斑东风螺3个选育世代遗传多样性和遗传结构的微卫星分析[J]. 海洋科学, 46(10): 85-93.
|
|
|
|
| [13] |
刘博, 邵艳卿, 王侃, 等, 2013. 4个缢蛏群体遗传多样性和系统发生关系的微卫星分析[J]. 海洋科学, 37(8): 96-102.
|
|
|
|
| [14] |
刘达博, 牛东红, 冯冰冰, 等, 2011. 乐清湾和三沙湾缢蛏群体遗传多样性的微卫星分析[J]. 上海海洋大学学报, 20(3): 350-357.
|
|
|
|
| [15] |
牛东红, 李家乐, 冯冰冰, 等, 2009. 缢蛏6个群体遗传结构的ISSR分析[J]. 应用与环境生物学报, 15(3): 332-336.
|
|
|
|
| [16] |
牛东红, 陈慧, 林国文, 等, 2010. 缢蛏群体微卫星分析中样本量对遗传多样性指标的影响[J]. 海洋科学进展, 28(2): 203-208.
|
|
|
|
| [17] |
牛东红, 冯冰冰, 刘达博, 等, 2011. 浙闽沿海缢蛏群体遗传结构的微卫星和线粒体CO Ⅰ序列分析[J]. 水产学报, 35(12): 1805-1813.
|
|
|
|
| [18] |
彭敏, 肖珊, 洪传远, 等, 2022. 华南沿海长鳍篮子鱼不同地理群体的遗传多样性分析[J]. 水生态学杂志, 43(5): 127-133.
|
|
|
|
| [19] |
阮惠婷, 徐姗楠, 李敏, 等, 2020. 飘鱼微卫星位点的筛选及珠江流域5个地理群体的遗传多样性分析[J]. 水生生物学报, 44(3): 501-508.
|
|
|
|
| [20] |
邵艳卿, 方军, 柏艳, 等, 2015. 缢蛏(Sinonovacula constricta) EST-SSR标记与生长性状的相关性分析[J]. 海洋与湖沼, 46(5): 1146-1152.
|
|
|
|
| [21] |
苏晓盈, 代永仙, 谭杰, 等, 2021. 马氏珠母贝(Pinctada martensii)黑壳色养殖群体SSR遗传多样性分析[J]. 基因组学与应用生物学, 40(2): 615-621.
|
|
|
|
| [22] |
孙志鹏, 鲁翠云, 那荣滨, 等, 2023. 利用线粒体序列比较分析梭鲈鸭绿江和乌伦古湖群体的遗传结构[J]. 水产学杂志, 36(5): 10-16, 26.
|
|
|
|
| [23] |
唐芳, 温贝妮, 刘红, 2021. 不同凡纳滨对虾养殖群体的微卫星遗传多样性分析[J]. 南方农业学报, 52(4): 1108-1115.
|
|
|
|
| [24] |
滕爽爽, 胡高宇, 范建勋, 等, 2021. 缢蛏5个群体遗传多样性和遗传分化的SNP分析[J]. 水生生物学报, 45(4): 861-870.
|
|
|
|
| [25] |
田镇, 陈爱华, 曹奕, 等, 2021. 红壳色文蛤选育群体遗传多样性的微卫星分析[J]. 南方农业学报, 52(9): 2582-2589.
|
|
|
|
| [26] |
王冬群, 李太武, 苏秀榕, 2005. 象山缢蛏养殖群体和野生群体遗传多样性的比较[J]. 中国水产科学, 12(2): 138-143.
|
|
|
|
| [27] |
吴玲, 2013. 马氏珠母贝和缢蛏的遗传多样性研究[D]. 青岛: 中国海洋大学: 49-86.
|
|
|
|
| [28] |
徐义平, 许会宾, 金凯, 等, 2017. 浙江乐清湾缢蛏的形态和遗传多样性[J]. 上海海洋大学学报, 26(1): 31-37.
|
|
|
|
| [29] |
于思梦, 高磊, 王伟, 等, 2017. 辽宁黄渤海沿岸长牡蛎遗传多样性分析[J]. 经济动物学报, 21(4): 215-220.
|
|
|
|
| [30] |
于颖, 孟祥盈, 王秀利, 等, 2007. 缢蛏遗传多样性的RAPD分析[J]. 生物技术通报, (6): 138-140.
|
|
|
|
| [31] |
张帝, 强俊, 傅建军, 等, 2022. 基于微卫星标记和线粒体D-loop序列的5个大口黑鲈群体遗传变异分析[J]. 中国水产科学, 29(9): 1277-1289.
|
|
|
|
| [32] |
张秀英, 张晓军, 赵翠, 等, 2012. 栉孔扇贝BES-SSR的开发及遗传多样性分析[J]. 水产学报, 36(6): 815-824.
|
|
|
|
| [33] |
赵文浩, 易少奎, 周琼, 等, 2023. 新疆塔里木河叶尔羌高原鳅群体遗传学研究[J]. 水产科学, 42(4): 664-673.
|
|
|
|
| [34] |
pmid: 6247908 |
| [35] |
|
| [36] |
|
| [37] |
doi: 10.1534/genetics.119.302370 pmid: 31537622 |
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
doi: 10.1126/science.3576198 pmid: 3576198 |
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
doi: 10.1016/j.margen.2012.06.003 pmid: 23904061 |
| [56] |
|
| [57] |
|
| [58] |
|
| [1] | HUANG Hongwei, ZHANG Zhixin, ZHONG Jia, LIN Qiang, GUO Baoying, YAN Xiaojun. The genetic structure and connectivity of eight fish species in the Indo-Pacific Convergence Region [J]. Journal of Tropical Oceanography, 2025, 44(1): 9-23. |
| [2] | HUANG Wen, FENG Yi, LI Ming, WU Qian, LUO Yanqiu, CHEN Yinmin, WANG Lirong, YU Kefu. A review on the population genetics of scleractinian corals [J]. Journal of Tropical Oceanography, 2024, 43(6): 13-26. |
| [3] | REN Huimin, ZHANG Heng, XU Shasha, LI Shengfa, LI Jiansheng, LI Zhihong, HE Lijun. Population genetic structure and historical dynamics of Trachurus japonicus in the China seas based on mitochondrial control region [J]. Journal of Tropical Oceanography, 2021, 40(5): 36-44. |
| [4] | ZOU Congcong, WANG Lijuan, WU Zhihao, YOU Feng. Population genetic structure of Japanese anchovy (Engraulis japonicus) in the Yellow Sea based on mitochondrial control region sequences* [J]. Journal of Tropical Oceanography, 2021, 40(5): 25-35. |
| [5] | Min LI, Xiaolan KONG, Youwei XU, Zuozhi CHEN. Genetic polymorphism of the Brushtooth lizardfish Saurida undosquamis based on mitochondrial D-loop sequences [J]. Journal of Tropical Oceanography, 2020, 39(4): 42-49. |
| [6] | Yingying JI,Lei XU,Hong LI,Lianggen WANG,Feiyan DU. Genetic structure of Oithona setigera from South China Sea based on 28S rDNA gene [J]. Journal of Tropical Oceanography, 2019, 38(3): 89-97. |
| [7] | Xiaolin HUANG, Wenjun LI, Heizhao LIN, Yukai YANG, Tao LI, Wei YU, Zhong HUANG. Genetic variations among Siganus oramin populations in coastal waters of southeast China based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2018, 37(4): 45-51. |
| [8] | Zhiying ZHAO, Liyun LIANG, Lirong BAI. Analysis of genetic diversity among three wild populations of Penaeus monodon using microsatellite marker [J]. Journal of Tropical Oceanography, 2018, 37(3): 65-72. |
| [9] | Yunchao DU, Shumei XIE, Shengyao HE, Donghong NIU, Jiale LI. Dopamine receptor genes of Sinonovacula constricta and its functions in damage healing of tissue [J]. Journal of Tropical Oceanography, 2018, 37(3): 45-54. |
| [10] | WENG Zhaohong, XIE Yangjie, XIAO Zhiqun, WANG Yilei. Analysis of genetic diversity in several wild and hatchery populations of Crassostrea angulata from south Fujian and south Guangdong [J]. Journal of Tropical Oceanography, 2016, 35(3): 94-98. |
| [11] | SHI Xiao-feng1, 2, SU Yong-quan1, WANG Wen-cheng1, WANG Jun1. Population genetic structure of three stocks of Acanthopagrus schlegelii based on mtDNA control region sequences [J]. Journal of Tropical Oceanography, 2015, 34(1): 56-63. |
| [12] | LIU Li, ZHAO Jie, GUO Yu-song, LIU Chu-wu. Study on the AFLP primer selection and genetic diversities of 4 Snappers [J]. Journal of Tropical Oceanography, 2012, 31(4): 112-116. |
| [13] | LI Li-hao,YU Da-hui,HUANG Gui-ju,DU Bo,FU Yun,TONG Xin,GUO Yi-hui,YE Wei. Comparison of genetic diversity among stocks of Oreochromis niloticus, O. aureus and red tilapia based on microsatellite DNA [J]. Journal of Tropical Oceanography, 2012, 31(2): 102-109. |
| [14] | JI Lei,OU You-jun,LI Jia-er. Genetic polymorphism of three cultured populations of golden pompano Trachinotus ovatus as revealed by microsatellites [J]. Journal of Tropical Oceanography, 2011, 30(3): 62-68. |
| [15] | GUO Yu-song,WANG Zhong-duo,LIU Chu-wu,CHEN Zhi-ming,LIU Yun. Isolation and genetic diversity analysis of microsatellites from nine species of familiar snappers [J]. Journal of Tropical Oceanography, 2010, 29(3): 82-86. |
|
||