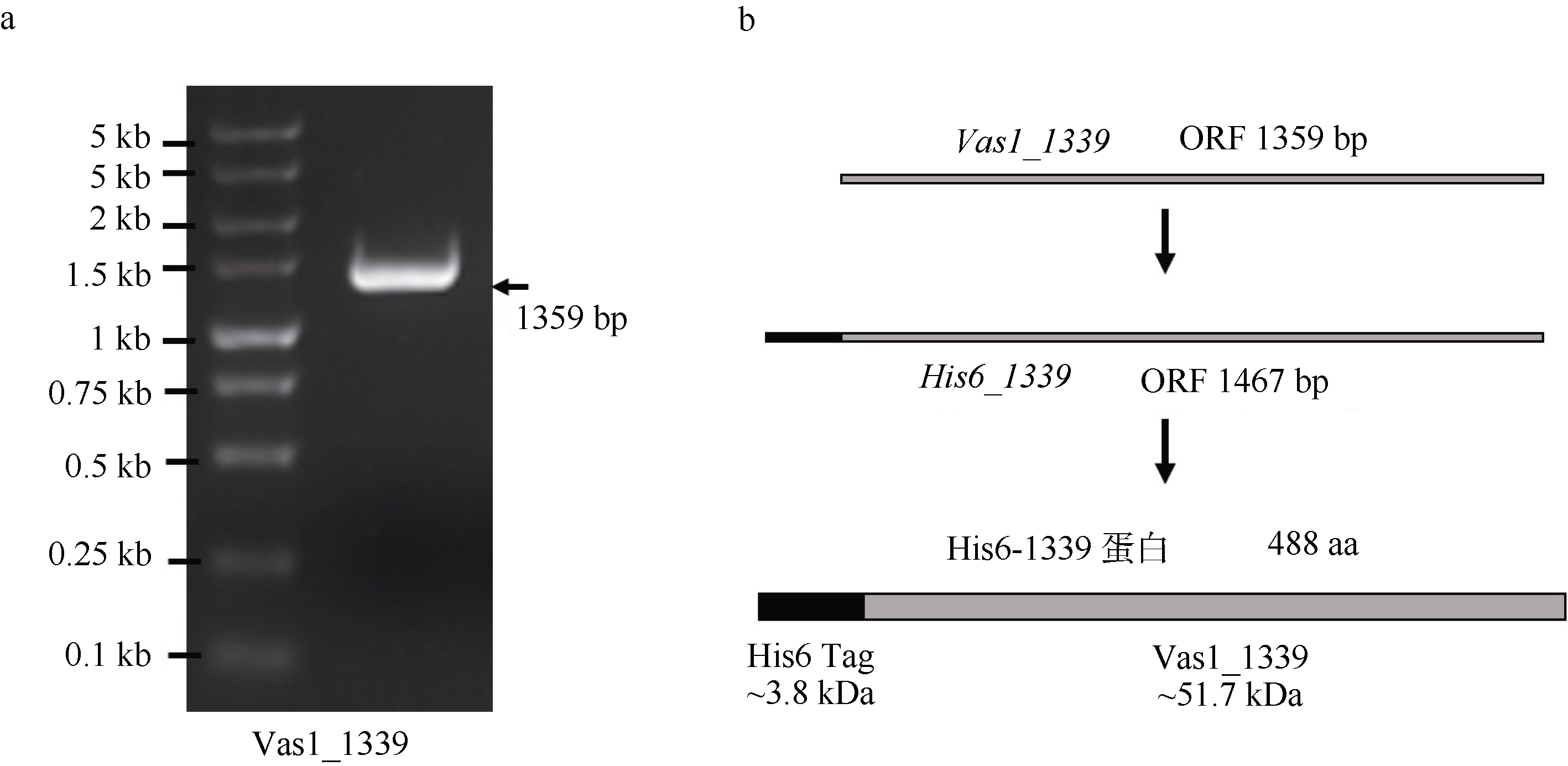
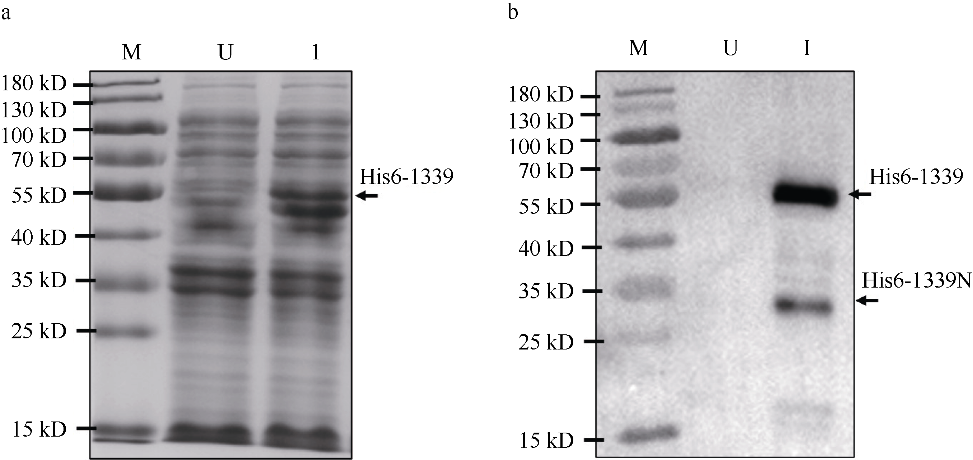
| [1] |
林福娣, 2020. 新琼寡糖的酶法制备及其生物活性研究[D]. 厦门: 华侨大学, 14-29.
|
|
LIN FUTI, 2020. Study on the enzymatic preparation and bioactivities of neoagaro- oligosaccharides[D]. Xiamen: Huaqiao University, 14-29. (in Chinese with English abstract)
|
| [2] |
张静雅, 刘宇鹏, 吴超, 等, 2018. 1株高产琼脂糖酶海洋弧菌的分离与酶活性的测定[J]. 热带生物学报, 9(2): 142-146.
|
|
ZHANG JINGYA, LIU YUPENG, WU CHAO, et al, 2018. Isolation and enzyme activity assay of an agarase-producing Vibrio sp. strain[J]. Journal of Tropical Biology, 9(2): 142-146. (in Chinese with English abstract)
|
| [3] |
ALKOTAINI B, HAN N S, KIM B S, 2016. Enhanced catalytic efficiency of endo-β-agarase Ⅰ by fusion of carbohydrate- binding modules for agar prehydrolysis[J]. Enzyme and Microbial Technology, 93-94: 142-149.
doi: 10.1016/j.enzmictec.2016.08.010
|
| [4] |
BARBEYRON T, GERARD A, POTIN P, et al, 1998. The kappa-carrageenase of the marine bacterium Cytophaga drobachiensis. Structural and phylogenetic relationships within family-16 glycoside hydrolases[J]. Molecular Biology and Evolution, 15(5): 528-537.
doi: 10.1093/oxfordjournals.molbev.a025952
|
| [5] |
BORASTON A, BOLAM D, GILBERT H, et al, 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition[J]. Biochemical Journal, 382(3): 769-781.
doi: 10.1042/BJ20040892
|
| [6] |
CHI W J, CHANG Y K, HONG S K, 2012. Agar degradation by microorganisms and agar-degrading enzymes[J]. Applied Microbiology and Biotechnology, 94(4): 917-930.
doi: 10.1007/s00253-012-4023-2
|
| [7] |
DONG JINHUA, TAMARU Y, ARAKI T, 2007. A unique β-agarase, AgaA, from a marine bacterium, Vibrio sp. strain PO-303[J]. Applied Microbiology and Biotechnology, 74(6): 1248-1255.
doi: 10.1007/s00253-006-0781-z
|
| [8] |
FU XIAOTING, KIM S M, 2010. Agarase: review of major sources, categories, purification method, enzyme characteristics and applications[J]. Marine Drugs, 8(1): 200-218.
doi: 10.3390/md8010200
|
| [9] |
HAN WENJUN, GU JINGYAN, LIU HUIHUI, et al, 2013. An extra peptide within the catalytic module of a β-Agarase affects the agarose degradation Pattern[J]. Journal of Biological Chemistry, 288(13): 9519-9531.
doi: 10.1074/jbc.M112.412247
|
| [10] |
HSU P H, WEI C H, LU W J, et al, 2015. Extracellular production of a novel endo-β-agarase AgaA from Pseudomonas vesicularis MA103 that cleaves agarose into neoagarotetraose and neoagarohexaose[J]. International Journal of Molecular Sciences, 16(3): 5590-5603.
doi: 10.3390/ijms16035590
|
| [11] |
JAM M, FLAMENT D, ALLOUCH J, et al, 2005. The endo-beta-agarases AgaA and AgaB from the marine bacterium Zobellia galactanivorans: two paralogue enzymes with different molecular organizations and catalytic behaviours[J]. Biochemical Journal, 385: 703-713.
doi: 10.1042/BJ20041044
|
| [12] |
JIANG CHENGCHENG, LIU ZHEN, CHENG DANYANG, et al, 2020. Agarose degradation for utilization: Enzymes, pathways, metabolic engineering methods and products[J]. Biotechnology Advances, 45: 107641.
doi: 10.1016/j.biotechadv.2020.107641
|
| [13] |
LIANG S S, CHEN Y P, CHEN Y H, et al, 2014. Characterization and overexpression of a novel β-agarase from Thalassomonas agarivorans[J]. Journal of Applied Microbiology, 116(3): 563-572.
doi: 10.1111/jam.12389
|
| [14] |
LIU YUPENG, JIN XINGKUN, WU CHAO, et al, 2020. Genome-Wide identification and functional characterization of β-Agarases in Vibrio astriarenae strain HN897[J]. Frontiers in Microbiology, 11: 1404.
doi: 10.3389/fmicb.2020.01404
|
| [15] |
LU XINZHI, CHU YAN, WU QIANQIAN, et al, 2009. Cloning, expression and characterization of a new agarase-encoding gene from marine Pseudoalteromonas sp.[J]. Biotechnology Letters, 31(10): 1565-1570.
doi: 10.1007/s10529-009-0042-1
|
| [16] |
MARTIN M, PORTETELLE D, MICHEL G, et al, 2014. Microorganisms living on macroalgae: diversity, interactions, and biotechnological applications[J]. Applied Microbiology and Biotechnology, 98(7): 2917-2935.
doi: 10.1007/s00253-014-5557-2
|
| [17] |
PARK S H, LEE C R, HONG S K, 2020. Implications of agar and agarase in industrial applications of sustainable marine biomass[J]. Applied Microbiology and Biotechnology, 104(7): 2815-2832.
doi: 10.1007/s00253-020-10412-6
|
| [18] |
POTIN P, RICHARD C, BARBEYRON T, et al, 1995. Processing and hydrolytic mechanism of the cgkA-Encoded κ- Carrageenase of Alteromonas carrageenovora[J]. European Journal of Biochemistry, 228(3): 971-975.
doi: 10.1111/ejb.1995.228.issue-3
|
| [19] |
TEH A H, FAZLI N H, FURUSAWA G, 2020. Crystal structure of a neoagarobiose-producing GH16 family β-agarase from Persicobacter sp. CCB-QB2[J]. Applied Microbiology and Biotechnology, 104(2): 633-641.
doi: 10.1007/s00253-019-10237-y
|
| [20] |
WANG HUA, ZHANG WEIBIN, CUI ZIBO, et al, 2020. Characterization of the hydrolysate and catalytic cavity of α- agarase AgaD[J]. Biotechnology Letters, 42(10): 1919-1925.
doi: 10.1007/s10529-020-02901-5
|
| [21] |
YU S, YUN E J, KIM D H, et al, 2020. Dual agarolytic pathways in a marine bacterium, Vibrio sp. strain EJY3: Molecular and enzymatic verification[J]. Applied and Environmental Microbiology, 86(6): e02724-19.
|
| [22] |
ZHANG WEIWEI, SUN LI, 2007. Cloning, characterization, and molecular application of a beta-agarase gene from Vibrio sp. strain V134[J]. Applied and Environmental Microbiology, 73(9): 2825-2831.
doi: 10.1128/AEM.02872-06
|
 ), JIN Xingkun, LEI Tianying, CAO Haihang, CHEN Qianghui, YANG Yaofan, LI Jiahang, ZHAO Zhe(
), JIN Xingkun, LEI Tianying, CAO Haihang, CHEN Qianghui, YANG Yaofan, LI Jiahang, ZHAO Zhe( )
)