Journal of Tropical Oceanography ›› 2025, Vol. 44 ›› Issue (1): 24-34.doi: 10.11978/2024067CSTR: 32234.14.2024067
• Marine Biology • Previous Articles Next Articles
Molecular cloning and functional study of cyclic GMP-AMP synthase from Crassostrea gigas
BAI Jing1,2( ), MAO Fan2, LIU Kelin2, SONG Jingchen2, YU Ziniu2, ZHANG Yang2(
), MAO Fan2, LIU Kelin2, SONG Jingchen2, YU Ziniu2, ZHANG Yang2( )
)
- 1. Jinan University, Guangzhou 510632, China
2. Key Laboratory of Tropical Marine Bio-resources and Ecology, South China Sea Institute of Oceanology, Chinese Academy of Science, Guangzhou 510301, China
-
Received:2024-03-22Revised:2024-04-10Online:2025-01-10Published:2025-02-10 -
Contact:ZHANG Yang -
Supported by:National Key Research and Development Program of China(2022YFD2400301); National Natural Science Foundation of China(32073002); National Natural Science Foundation of China(U22A20533); 9th Young Elite Scientists Sponsorship Program(2023QNRC001); Science and Technology Planning Project of Guangdong Province, China(2023A04J0096); Science and Technology Planning Project of Guangdong Province, China(2024A04J6278)
CLC Number:
- P735
Cite this article
BAI Jing, MAO Fan, LIU Kelin, SONG Jingchen, YU Ziniu, ZHANG Yang. Molecular cloning and functional study of cyclic GMP-AMP synthase from Crassostrea gigas[J].Journal of Tropical Oceanography, 2025, 44(1): 24-34.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab. 1
Sequences of designed primers used in this study"
| 引物名称 | 序列(5′-3′) | 用途 |
|---|---|---|
| cGAS-F1 | ATGGTAATCAAATGTCCTAATTGTG | ORF克隆 |
| cGAS-R1 | TTATTGTAAGAGACCCTCTAGTTCC | ORF克隆 |
| pEGFP-N1-cGAS-F | TCAGATCTCGAGCTCAAGCTTGCCACCATGGTAATCAAATGTCC | pEGFP-N1-cGAS重组质粒 |
| pEGFP-N1-cGAS-R | ATGGTGGCGACCGGTGGATCCGATTGTAAGAGACCCTCTAGTTCCCT | pEGFP-N1-cGAS重组质粒 |
| PCDNA3.1/V5-His-cGAS-F | GCACAGTGGCGGCCGCTCGAGATGGTAATCAAATGTCCTAATTGTGAC | pcDNA3.1/V5-His-cGAS重组质粒 |
| PCDNA3.1/V5-His-cGAS-R | AGGCTTACCTTCGAAGGGCCCTTGTAAGAGACCCTCTAGTTCCCTG | pcDNA3.1/V5-His-cGAS重组质粒 |
| dscGAS-F | CGACAAAACAAAGATCGACTACAAC | cGAS-RNAi |
| dscGAS-R | CAGTCTGGATTTCTCTTGCATCCTT | cGAS-RNAi |
| dscGAS-F-T7 | TAATACGACTCACTATAGGCGACAAAACAAAGATCGACTACAAC | cGAS-RNAi |
| dscGAS-R-T7 | TAATACGACTCACTATAGGCAGTCTGGATTTCTCTTGCATCCTT | cGAS-RNAi |
| dsGFP-F | GCAAGGGCGAGGAGCTGTTCACCGG | GFP-RNAi |
| dsGFP-R | TTGCCGTCCTCCTTGAAGTCGATGC | GFP-RNAi |
| dsGFP-F-T7 | TAATACGACTCACTATAGGGCAAGGGCGAGGAGCTGTTCACCGG | GFP-RNAi |
| dsGFP-R-T7 | TAATACGACTCACTATAGGTTGCCGTCCTCCTTGAAGTCGATGC | GFP-RNAi |
| cGAS-F2 | GGAAAGACGACAGGGACGG | qRT-PCR |
| cGAS-R2 | TGTCTGGAGAACCCCTTTGG | qRT-PCR |
| viperin-F | CTGAAACCCATCAGTGTCAACTACC | qRT-PCR |
| viperin-R | GACAATGAAGGGCTCGCCAC | qRT-PCR |
| IRF2-F | ACTTCCGCTGTGCCCTGAAT | qRT-PCR |
| IRF2-R | TATGACCTTTGGCACTGTCGTTC | qRT-PCR |
| IL-17-F | AAACATGCTGGAATACCTCGAGT | qRT-PCR |
| IL-17-R | GTGGGACGCTACGAGGAAATA | qRT-PCR |
| TNF-F | GTATTACTGAGGAATGGCGTGAAAC | qRT-PCR |
| TNF-R | CAAAATCCCTGTCACTACAACCG | qRT-PCR |
| IRF8-F | GGACAGCGGTCAGACACGAC | qRT-PCR |
| IRF8-R | CCTTGAATATTGTGGAGTCTGCCT | qRT-PCR |
| β-actin-F | GGATTGGCGTGGTGGTAGAG | qRT-PCR |
| β-actin-R | GTATGATGCCCCTTTGTTGAGTC | qRT-PCR |
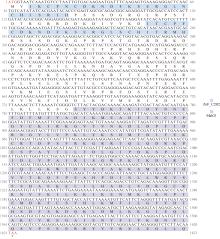
Fig. 1
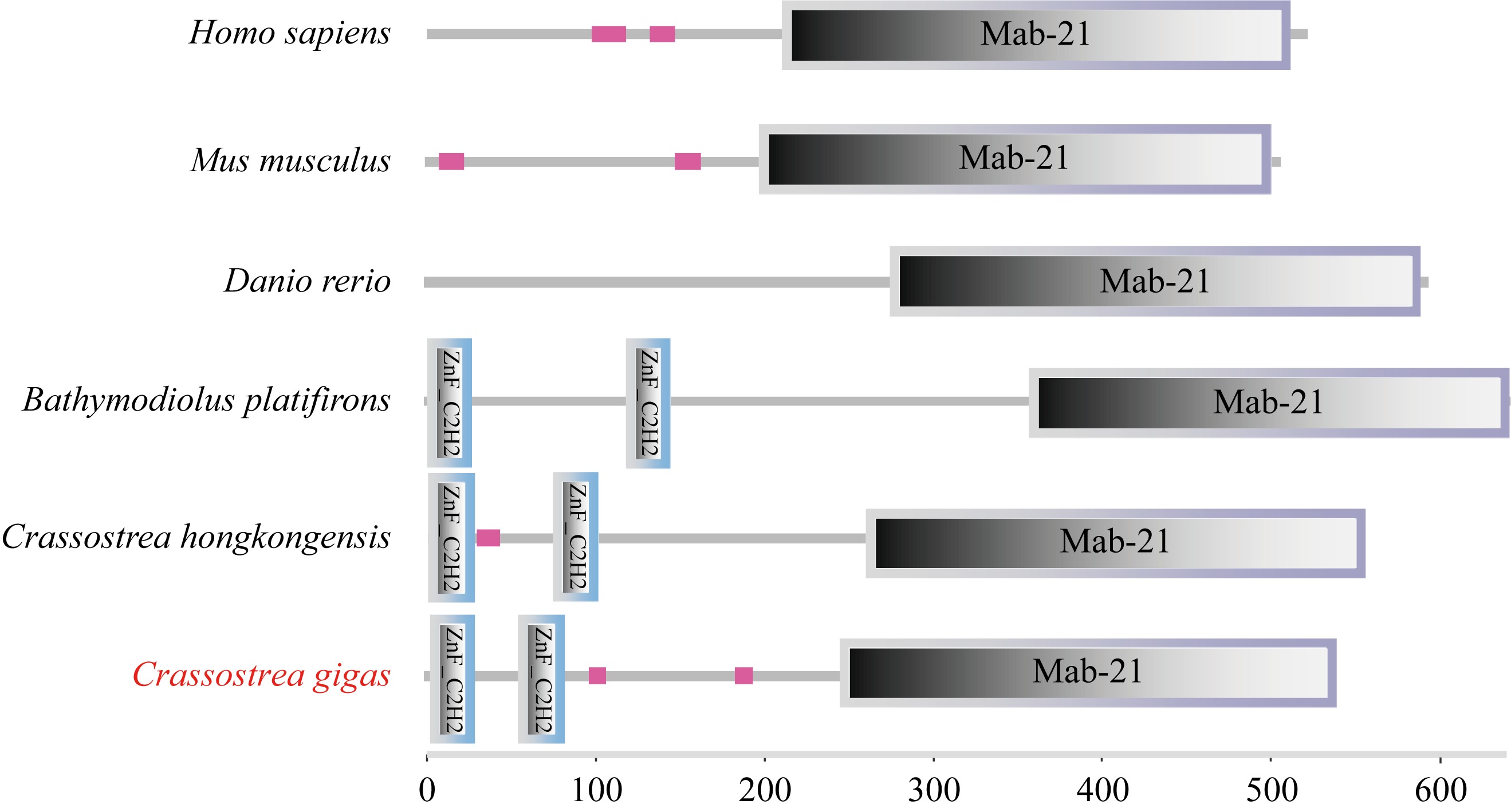
Full length of the Crassostrea gigas cGAS open reading frame sequence and deduced amino acid sequence. The zinc finger structure is marked with a blue box and the Mab21 structural domain is marked with a purple box. * indicates protein translation termination, with initiation codon ATG and termination codon TAA in red font"


Fig. 3
Multiple alignment of cGAS amino acid sequences between Crassostrea gigas and other species. NTase catalytical sites are marked with red dots, and the zinc-ribbon domain is indicated by a box, with “---” representing an amino acid deletion. “*”, “:” and “.” present identical, highly conserved, and less conserved amino acid residues, respectively"

Fig. 6
Subcellular localization of cGAS in Crassostrea gigas. Shown on the left is the fluorescent fusion protein of pEGFP and cGAS; shown in the middle is the nucleus after DAPI staining; and shown on the right is the combined image. The microscopic observation results of HEK293T cells at 40 magnifications"
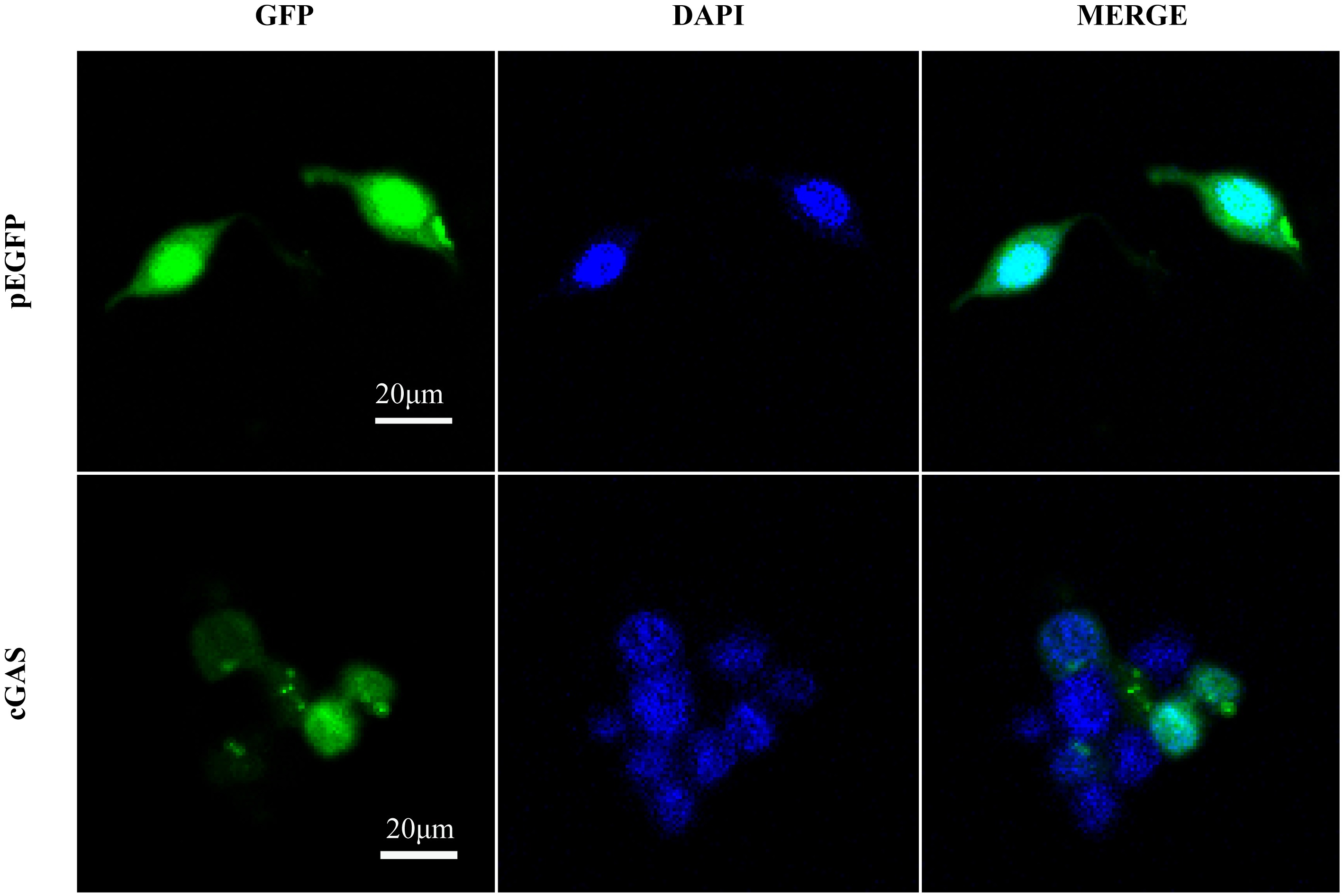

Fig. 8
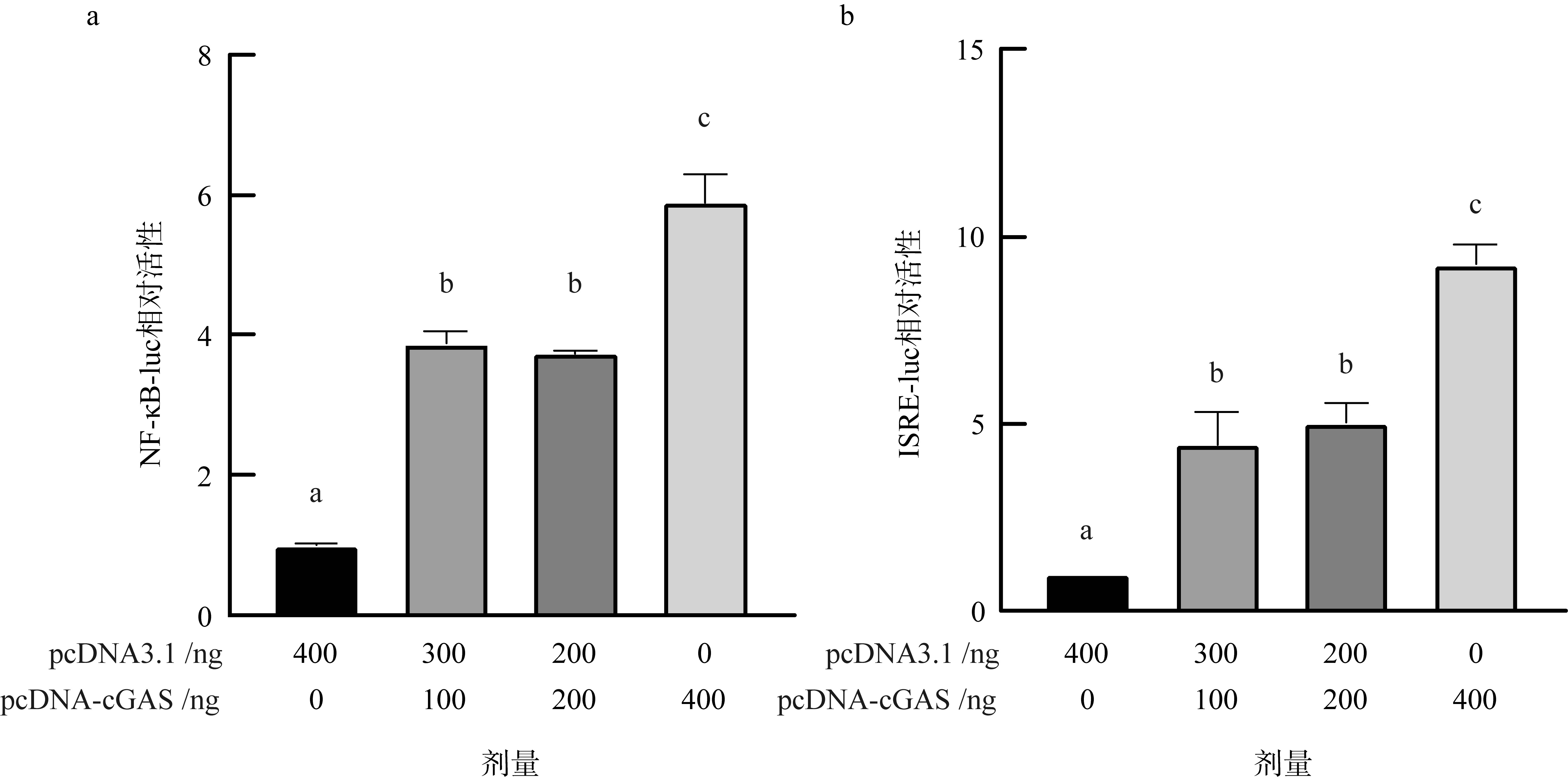
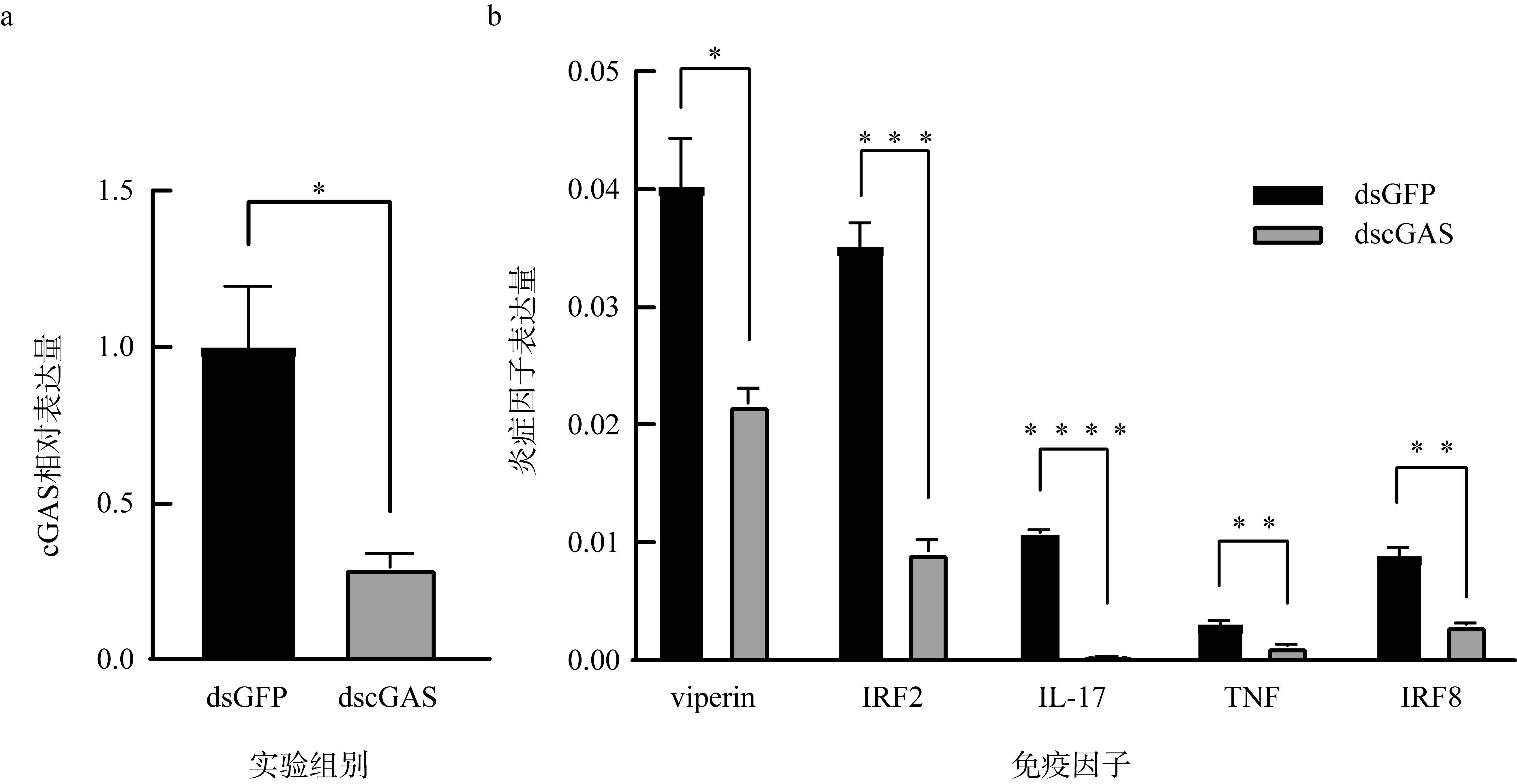
Expression of downstream factors after cGAS knockdown in Crassostrea gigas; (a) fluorescence quantitative PCR was used to detect the knockdown effect; (b) expression of downstream immune factors after cGAS knockdown. All data are presented as mean ± SEM, with significance denoted as * for P<0.05, ** for P<0.01, *** for P<0.001, and **** for P<0.0001"
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [1] | JIANG Rouyun, JIAN Lili, SHI Songbiao, TIAN Xinpeng. Diversity of culturable bacteria in the sedimentary sands of the Meiji Reef [J]. Journal of Tropical Oceanography, 2024, 43(6): 170-180. |
| [2] | RAO Yiyong, ZHAO Meirong, KUANG Zexing, HUANG Honghui, TAN Erhui. Influence of raft-string oyster culture on the functional structure of macrobenthic communities: a case study in the Dapeng Cove* [J]. Journal of Tropical Oceanography, 2024, 43(5): 69-83. |
| [3] | JIA Nan, ZHOU Tiancheng, HU Simin, ZHANG Chen, HUANG Hui, LIU Sheng. Difference in the feeding contents of three hermit crabs in the coral reefs of the Nansha Islands, South China Sea [J]. Journal of Tropical Oceanography, 2024, 43(3): 109-121. |
| [4] | LI Cai, LIU Cong, ZHANG Xianqing, CHEN Fei, XIAO Zhihui, YANG Zeming, ZHENG Yuanning, ZHOU Wen, XU Zhantang. Development and Application of the Multiangle Volume Scattering and Attenuation Meter (VSAM)* [J]. Journal of Tropical Oceanography, 2024, 43(2): 1-11. |
| [5] | LI Ze, WANG Lijuan, ZOU Congcong, SHU Chang, WU Zhihao, ZOU Yuxia, YOU Feng. Expression characteristics and function of Anti-Müllerian hormone receptor Ⅱ gene (amhr2) in Paralichthys olivaceus [J]. Journal of Tropical Oceanography, 2024, 43(1): 94-106. |
| [6] | SUN Tingting, HAO Wenjin, XU Pengzhen, YE Lijing, DONG Zhijun. Effects of seawater acidification on microorganisms associated with Aurelia coerulea polyps [J]. Journal of Tropical Oceanography, 2023, 42(6): 111-119. |
| [7] | ZHANG Xianqing, LI Cai, Zhou Wen, LIU Cong, XU Zhantang, CAO Wenxi, YANG Yuezhong. Studying on diffuse attenuation coefficient in the South China Sea based on volume scattering function and absorption coefficient* [J]. Journal of Tropical Oceanography, 2023, 42(3): 86-95. |
| [8] | TANG Chaoli, TAO Xinhua, WEI Yuanyuan, DAI Congming, WEI Heli. Spatiotemporal modal analysis and prediction of surface temperature in East Asia and the Western Pacific* [J]. Journal of Tropical Oceanography, 2022, 41(6): 183-192. |
| [9] | XIE Yong, WANG Youshao, ZHANG Weishi. Cloning and expression analysis of Cinnamate-4-hydroxylase gene from Rhizophora stylosa [J]. Journal of Tropical Oceanography, 2022, 41(6): 20-27. |
| [10] | FENG Bingbin, WANG Riming, LI Shushi, HUANG Hu, HU Baoqing. Changes of the artificial beach profile in the Qinzhou Bay [J]. Journal of Tropical Oceanography, 2022, 41(4): 51-60. |
| [11] | CHEN Kehai, XIE Xuetong, ZHANG Jinlan, ZHENG Yan. An SST dependent geophysical model function for HY-2A scatterometer [J]. Journal of Tropical Oceanography, 2022, 41(2): 90-102. |
| [12] | LI Ao, FENG Yang, WANG Yuntao, XUE Huijie. Spatiotemporal variation of water area with high chlorophyll a concentration in the South China Sea based on OC-CCI data* [J]. Journal of Tropical Oceanography, 2022, 41(2): 77-89. |
| [13] | ZHAO Zehui, ZHANG Aijiao, YANG Yucheng, MAO Fan, XIAO Shu, LI Jun, ZHANG Yang, XIANG Zhiming, YU Ziniu. Molecular cloning and functional studies of ChPerlucin in Crassostrea hongkongensis [J]. Journal of Tropical Oceanography, 2022, 41(1): 42-51. |
| [14] | ZHANG Xiangyu, SONG Jingchen, LIU Kunna, MAO Fan, XIAO Shu, XIANG Zhiming, ZHANG Yang, YU Ziniu. Cloning and functional analysis of a new pattern recognition receptor CgLRRC69 in Pacific oyster, Crassostrea gigas [J]. Journal of Tropical Oceanography, 2021, 40(1): 65-74. |
| [15] | Shibing ZHU,Danni HU,Huiling ZHANG,Chunhua ZENG,Zhehua LI,Zhiqiang LI. Analysis of short-term temporal and spatial changes and sedimentary dynamics at the middle section of Haikou Bay Beach * [J]. Journal of Tropical Oceanography, 2019, 38(5): 77-85. |
|
||