Journal of Tropical Oceanography ›› 2025, Vol. 44 ›› Issue (1): 133-145.doi: 10.11978/2024059CSTR: 32234.14.2024059
• Exploitation of Marine Resources • Previous Articles Next Articles
Microbial diversity of potential EHMC-degrading bacteria from coral
HUANG Qinyu1,2( ), LYU Lina2, LI Jie2, JU Huimin2, SU Hongfei1(
), LYU Lina2, LI Jie2, JU Huimin2, SU Hongfei1( )
)
- 1. School of Marine Sciences, Guangxi University, Nanning 530004, China
2. Key Laboratory of Tropical Marine Bio-resources and Ecology, Chinese Academy of Sciences, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou 510301, China
-
Received:2024-03-18Revised:2024-04-12Online:2025-01-10Published:2025-02-10 -
Contact:SU Hongfei -
Supported by:National Natural Science Foundation of China(U23A2036)
CLC Number:
- P745
Cite this article
HUANG Qinyu, LYU Lina, LI Jie, JU Huimin, SU Hongfei. Microbial diversity of potential EHMC-degrading bacteria from coral[J].Journal of Tropical Oceanography, 2025, 44(1): 133-145.
share this article
Add to citation manager EndNote|Reference Manager|ProCite|BibTeX|RefWorks
Tab.1
The information of coral species collected from Luhuitou, Sanya"
| 珊瑚属名 | 珊瑚种名 | 拉丁名 |
|---|---|---|
| 蔷薇珊瑚属 | 截顶蔷薇珊瑚 | Montipora truncata |
| 蔷薇珊瑚 | Montipora sp. | |
| 盘星珊瑚属 | 盘星珊瑚 | Dipsastraea sp. |
| 牡丹珊瑚属 | 厚板牡丹珊瑚 | Pavona duerdeni |
| 十字牡丹珊瑚 | Pavona decussata | |
| 鹿角珊瑚科属 | 巨锥鹿角珊瑚 | Acropora monticulosa |
| 多孔鹿角珊瑚 | Acropora millepora | |
| 风信子鹿角珊瑚 | Acropora hyacinthus | |
| 鹿角珊瑚 | Acropora sp. | |
| 盔形珊瑚属 | 从生盔珊瑚 | Galaxea fascicularis |
| 角孔珊瑚属 | 角孔珊瑚 | Goniopora sp. |
| 角蜂巢珊瑚属 | 角蜂巢珊瑚 | Favosites sp. |
| 滨珊瑚属 | 澄黄滨珊瑚 | Porites lutea |
| 杯形珊瑚属 | 鹿角杯形珊瑚 | Pocillopora damicornis |
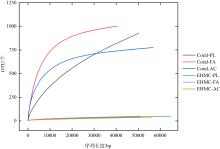
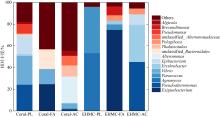
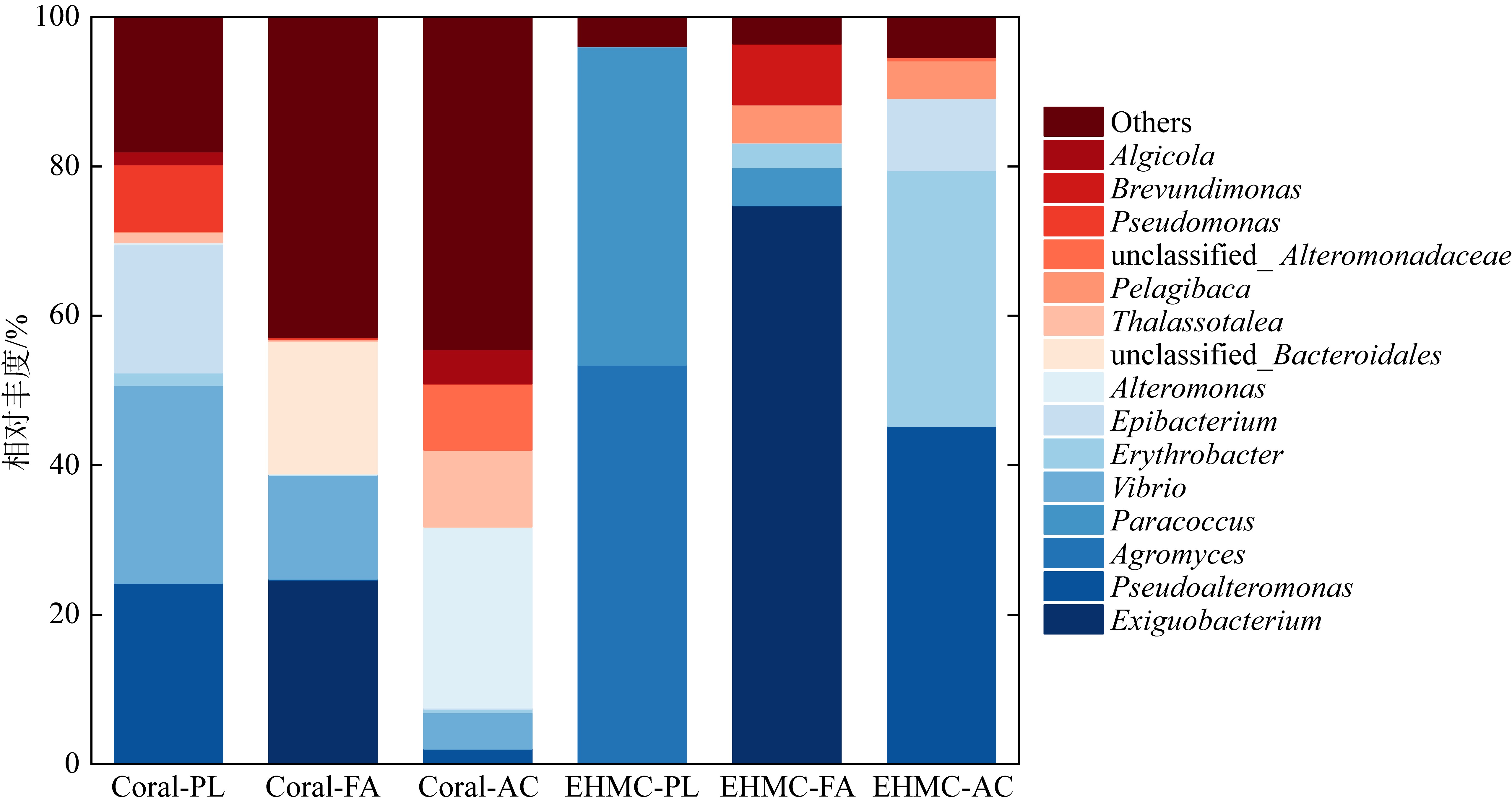
Tab. 2
Diversity indexes of microbial communities isolated from original coral and EHMC-enrichment samples"
| 珊瑚物种 | 样品编号 | ACE指数 | Chao1指数 | Simpson 指数 | Shannon 指数 |
|---|---|---|---|---|---|
| 滨珊瑚 | Coral-PL | 1888 | 1423 | 0.84 | 3.88 |
| EHMC-PL | 75 | 95 | 0.53 | 1.34 | |
| 角蜂巢珊瑚 | Coral-FA | 1040 | 1052 | 0.90 | 5.45 |
| EHMC-FA | 84 | 69 | 0.43 | 1.53 | |
| 鹿角珊瑚 | Coral-AC | 808 | 837 | 0.93 | 5.92 |
| EHMC-AC | 82 | 81 | 0.67 | 1.98 |
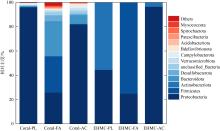
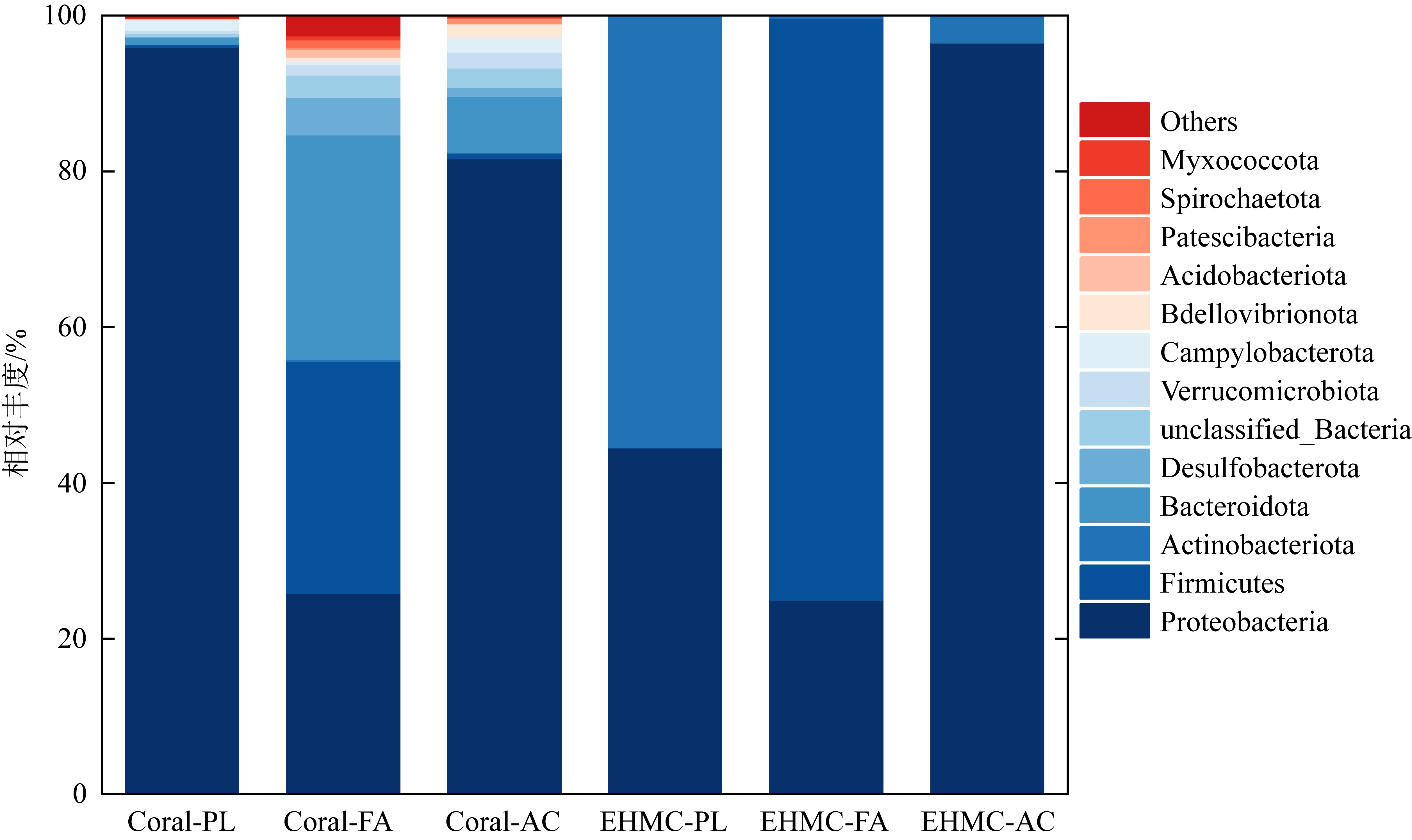
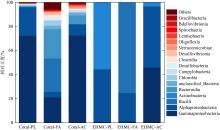
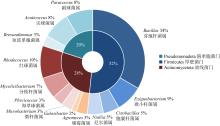
Tab. 3
The numbers of bacteria species at different levels in original coral and EHMC-enrichment samples"
| 珊瑚物种 | 样品编号 | 门数目 | 纲数目 | 目数目 | 科数目 | 属数目 |
|---|---|---|---|---|---|---|
| 滨珊瑚 | Coral-PL | 26 | 48 | 118 | 193 | 335 |
| EHMC-PL | 5 | 7 | 21 | 33 | 43 | |
| 角蜂巢珊瑚 | Coral-FA | 31 | 64 | 154 | 250 | 394 |
| EHMC-FA | 6 | 8 | 20 | 31 | 40 | |
| 鹿角珊瑚 | Coral-AC | 19 | 38 | 99 | 174 | 346 |
| EHMC-AC | 4 | 6 | 16 | 25 | 37 |
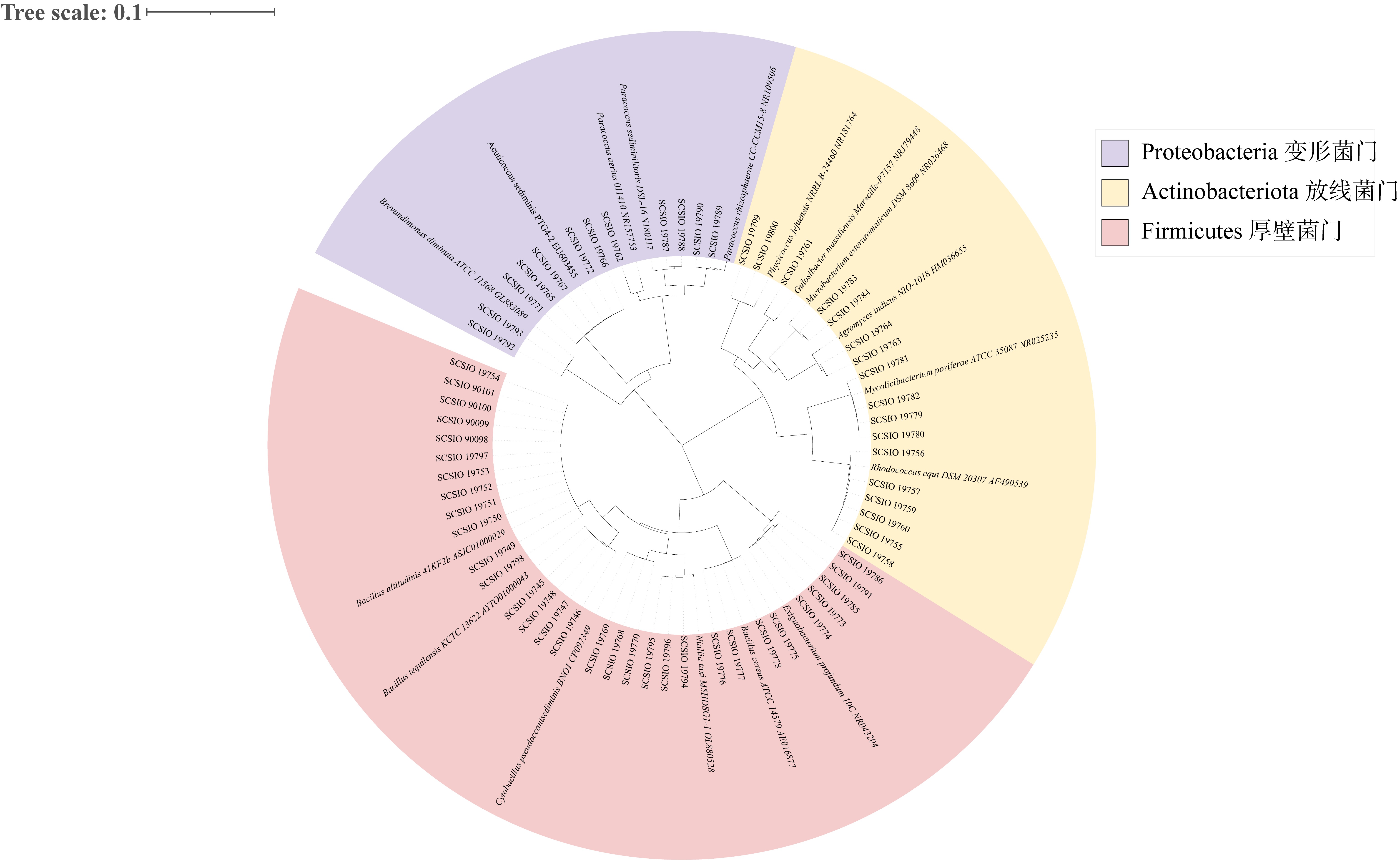
Tab. 4
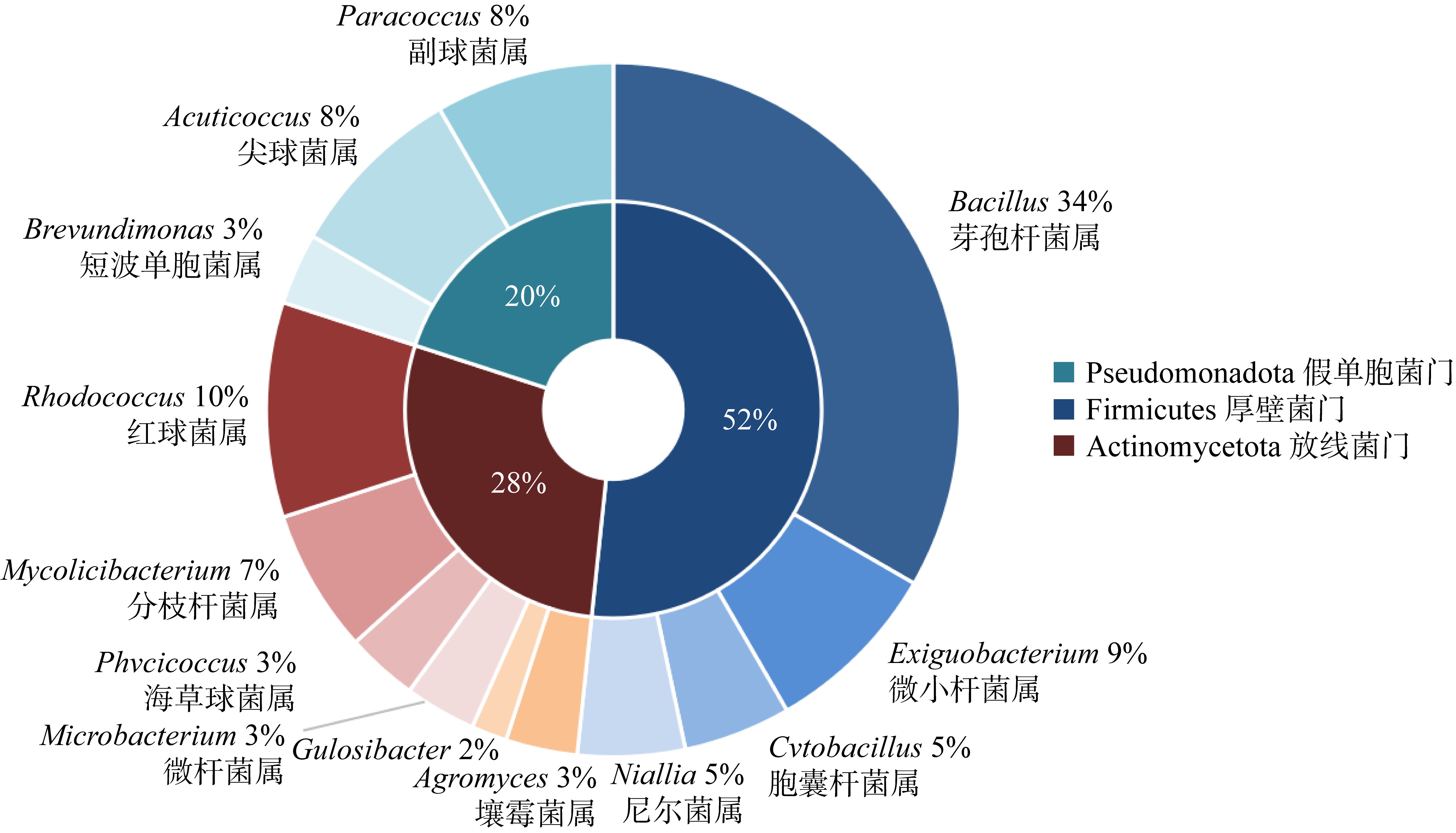
The information of potential EHMC-degrading strains"
| 菌株来源 | 菌株编号 | 种名 | 相似度/% |
|---|---|---|---|
| 杯形珊瑚 | SCSIO 19749 | Bacillus altitudinis 41KF2b | 100.00 |
| 角蜂巢珊瑚 | SCSIO 19792 | Brevundimonas diminuta ATCC 11568 | 99.77 |
| 角蜂巢珊瑚 | SCSIO 19789 | Paracoccus rhizosphaerae CC-CCM15-8 | 99.17 |
| 鹿角珊瑚 | SCSIO 19765 | Acuticoccus sediminis PTG4-2 | 100.00 |
| 鹿角珊瑚 | SCSIO 19799 | Phycicoccus jejuensis NRRL B-24460 | 99.13 |
| 鹿角珊瑚 | SCSIO 19755 | Rhodococcus hoagii DSM 20295 | 99.42 |
| 牡丹珊瑚 | SCSIO 19779 | Mycolicibacterium poriferae ATCC 35087 | 99.86 |
| 滨珊瑚 | SCSIO 19762 | Paracoccus aerius 011410 | 97.59 |
| [1] |
陈飚, 2021. 南海珊瑚微生物组的空间变化及其环境适应机制[D]. 南宁: 广西大学.
|
|
|
|
| [2] |
崔倩倩, 刘朝阳, 2020. 石油烃类污染物的微生物修复研究进展[J]. 江西科学, 38(3): 326-330, 384.
|
|
|
|
| [3] |
李明, 2022. 南海广布种澄黄滨珊瑚和低纬度优势种疣状杯形珊瑚的群体遗传学研究[D]. 南宁: 广西大学.
|
|
|
|
| [4] |
李汶璐, 王志超, 杨文焕, 等, 2022. 微塑料对沉积物细菌群落组成和多样性的影响[J]. 环境科学, 43(5): 2606-2613.
|
|
|
|
| [5] |
钱程, 2022. 新型二苯甲酮-3脱毒降解菌Pandoraea pnomenusa的筛选及降解特性研究[D]. 上海: 上海师范大学.
|
|
|
|
| [6] |
田蕴, 郑天凌, 2004. 海洋环境中降解多环芳烃的微生物[J]. 海洋科学, 28(9): 50-55.
|
|
|
|
| [7] |
王任, 吴鸳鸯, 程巧鸳, 2023. 2021年市售防晒类化妆品中防晒剂使用情况分析[J]. 香料香精化妆品, (1): 35-41.
|
|
|
|
| [8] |
吴钟解, 陈石泉, 陈敏, 等, 2013. 海南岛造礁石珊瑚资源初步调查与分析[J]. 海洋湖沼通报, (2): 44-50.
|
|
|
|
| [9] |
赵美霞, 余克服, 张乔民, 等, 2008. 三亚鹿回头石珊瑚物种多样性的空间分布[J]. 生态学报, 28(4): 1419-1428.
|
|
|
|
| [10] |
周楠, 姜成英, 刘双江, 2016. 从环境中分离培养微生物: 培养基营养水平至关重要[J]. 微生物学通报, 43(5): 1075-1081.
|
|
|
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [1] | HE Yueming, ZHAO Lining, CHEN Xinqi, HE Jiahong, FAN Hao, CHEN Leyi, ZHANG Cuixian, HE Xixin. γ-Aromatic butenolides lignans from soft coral-associated symbiotic and epiphytic fungi Aspergillus terreus EGF7-0-1(I) [J]. Journal of Tropical Oceanography, 2025, 44(1): 146-153. |
| [2] | QIU Xingyu, LIU Qingxia, CHEN Zuozhi, CAI Yancong, HUANG Honghui. Characteristics of carbon and nitrogen stable isotopes of major fish species in coral reefs of the Nansha Islands in spring 2023 [J]. Journal of Tropical Oceanography, 2024, 43(6): 104-113. |
| [3] | HUANG Wen, FENG Yi, LI Ming, WU Qian, LUO Yanqiu, CHEN Yinmin, WANG Lirong, YU Kefu. A review on the population genetics of scleractinian corals [J]. Journal of Tropical Oceanography, 2024, 43(6): 13-26. |
| [4] | JIANG Rouyun, JIAN Lili, SHI Songbiao, TIAN Xinpeng. Diversity of culturable bacteria in the sedimentary sands of the Meiji Reef [J]. Journal of Tropical Oceanography, 2024, 43(6): 170-180. |
| [5] | XIE Hongyu, LIU Yong, LI Chunhou, ZHAO Jinfa, SUN Jinhui, SHEN Jianzhong, SHI Juan, WANG Teng. Species composition and evolutionary characteristics of coral reef fish in the Langhua Reef, Xisha Islands [J]. Journal of Tropical Oceanography, 2024, 43(6): 114-128. |
| [6] | HUANG Liangmin, LIN Qiang, TAN Yehui, HUANG Xiaoping, ZHOU Linbin, HUANG Hui. Thoughts on the restoration, reconstruction and protection of typical tropical marine ecosystems [J]. Journal of Tropical Oceanography, 2024, 43(6): 1-12. |
| [7] | SUN Manman, ZENG Yanbo, XU Han, YAO Ligong, GUO Yuewei, SU Mingzhi. Chemical composition and antibacterial activities of the soft coral Lobophytum sp. from the South China Sea [J]. Journal of Tropical Oceanography, 2024, 43(4): 189-197. |
| [8] | MO Danyang, NING Zhiming, YANG Bin, XIA Ronglin, LIU Zhijin. Response of dissimilatory nitrate reduction processes in coral reef sediments of the Weizhou island to temperature changes [J]. Journal of Tropical Oceanography, 2024, 43(4): 137-143. |
| [9] | JIANG Lyumiao, CHEN Tianran, ZHAO Kuan, ZHANG Ting, XU Lijia. Experimental study on bioerosion of marginal reefs in the Weizhou Island, northern South China Sea [J]. Journal of Tropical Oceanography, 2024, 43(3): 155-165. |
| [10] | JIA Nan, ZHOU Tiancheng, HU Simin, ZHANG Chen, HUANG Hui, LIU Sheng. Difference in the feeding contents of three hermit crabs in the coral reefs of the Nansha Islands, South China Sea [J]. Journal of Tropical Oceanography, 2024, 43(3): 109-121. |
| [11] | PENG Erman, YAO Yu, LI Zhuangzhi, XU Conghao. Numerical study of the hydrodynamic characteristics of reef coast under the combined effects of waves and currents [J]. Journal of Tropical Oceanography, 2024, 43(3): 187-194. |
| [12] | LUO Yong, HUANG Lintao, YANG Jianhui, LIAN Jiansheng, LIU Chengyue, JIANG Lei, LIANG Yuxian, CHEN Lunju, LEI Xinming, LIU Sheng, HUANG Hui. Community structure of reef-building corals and their environmental impact factors in the coastal waters of Hongpai-Maniao, Lingao, Hainan [J]. Journal of Tropical Oceanography, 2024, 43(3): 72-86. |
| [13] | HUANG Hui, YUAN Xiangcheng, SONG Yan, LI Yingxin, ZHOU Weihua, LONG Aimin. Carbon sequestration process and carbon storage mechanism of reef ecosystem in South China Sea* [J]. Journal of Tropical Oceanography, 2024, 43(3): 13-21. |
| [14] | WANG Yongzhi, XU Lijia, HUANG Baiqiang, YANG Tianjian, QI Shibin, CHEN Hui, YANG Jing. Physiological responses to light limitation of reef-building corals in the Yongle Atoll of the Xisha Islands [J]. Journal of Tropical Oceanography, 2024, 43(3): 31-39. |
| [15] | HUANG Hui, YU Xiaolei, HUANG Lintao, JIANG Lei. Current status and prospects of coral reef ecology research [J]. Journal of Tropical Oceanography, 2024, 43(3): 3-12. |
|
||